
Persephonella marina (strain DSM 14350 / EX-H1)
Taxonomy: cellular organisms; Bacteria; Aquificae; Aquificae; Aquificales; Hydrogenothermaceae; Persephonella; Persephonella marina
Average proteome isoelectric point is 6.82
Get precalculated fractions of proteins
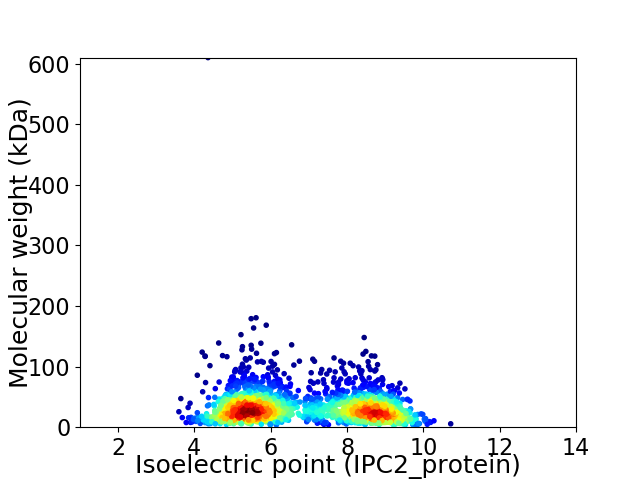
Virtual 2D-PAGE plot for 2048 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|C0QRJ0|C0QRJ0_PERMH Protoglobin domain-containing protein OS=Persephonella marina (strain DSM 14350 / EX-H1) OX=123214 GN=PERMA_1519 PE=4 SV=1
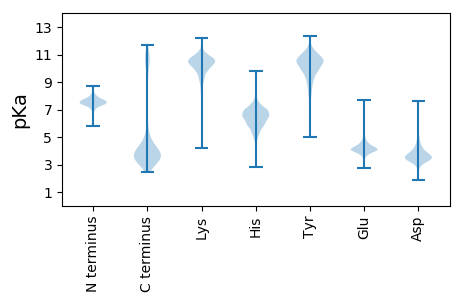
MM1 pKa = 7.61RR2 pKa = 11.84KK3 pKa = 8.96EE4 pKa = 4.15FKK6 pKa = 10.4KK7 pKa = 10.61KK8 pKa = 10.53AILGGSLSLSALLFACGGAGDD29 pKa = 4.14VASDD33 pKa = 3.58SPYY36 pKa = 9.86ATVTGTVTSASSTGSLSTTGIDD58 pKa = 2.89IGNVNLSFIAAVAIDD73 pKa = 3.94NGKK76 pKa = 9.49LVYY79 pKa = 9.44TADD82 pKa = 5.55DD83 pKa = 3.52IDD85 pKa = 4.3NNGNFNLKK93 pKa = 10.22LKK95 pKa = 10.51EE96 pKa = 4.08GLDD99 pKa = 3.54YY100 pKa = 11.57AFILFDD106 pKa = 3.35ISKK109 pKa = 9.9KK110 pKa = 9.99PKK112 pKa = 10.55LIVKK116 pKa = 9.75DD117 pKa = 3.89SNGNAIKK124 pKa = 10.59INGDD128 pKa = 3.12GSVNISLSDD137 pKa = 3.5SDD139 pKa = 4.84GDD141 pKa = 4.59GIPDD145 pKa = 3.67TASVNITGSVSLTKK159 pKa = 10.54DD160 pKa = 3.08SQLDD164 pKa = 3.84DD165 pKa = 4.41NDD167 pKa = 3.5NDD169 pKa = 4.29HH170 pKa = 6.7YY171 pKa = 11.23PDD173 pKa = 4.85GIEE176 pKa = 4.54NINAGGSVNPDD187 pKa = 2.98YY188 pKa = 11.6DD189 pKa = 3.83EE190 pKa = 6.35DD191 pKa = 5.49GDD193 pKa = 4.73GIFDD197 pKa = 5.24GIEE200 pKa = 4.13DD201 pKa = 4.34KK202 pKa = 11.5DD203 pKa = 3.72GDD205 pKa = 4.26GYY207 pKa = 10.71IDD209 pKa = 3.45GHH211 pKa = 6.31EE212 pKa = 4.67DD213 pKa = 3.45SNSNGIPDD221 pKa = 4.07VYY223 pKa = 11.1EE224 pKa = 5.72DD225 pKa = 4.62DD226 pKa = 5.92DD227 pKa = 6.15KK228 pKa = 12.02DD229 pKa = 3.8GLPNYY234 pKa = 10.38LDD236 pKa = 5.29DD237 pKa = 5.29SDD239 pKa = 5.27GDD241 pKa = 4.19GYY243 pKa = 9.96PDD245 pKa = 5.8HH246 pKa = 8.06IDD248 pKa = 3.64EE249 pKa = 5.8DD250 pKa = 4.79DD251 pKa = 3.76SDD253 pKa = 4.27GYY255 pKa = 10.66KK256 pKa = 10.28YY257 pKa = 10.79EE258 pKa = 4.16MKK260 pKa = 10.06GTVSNIDD267 pKa = 3.34TTTGTFVFTYY277 pKa = 10.58NGTDD281 pKa = 3.47YY282 pKa = 11.0NVSVTDD288 pKa = 3.53TTVCEE293 pKa = 4.46INDD296 pKa = 4.03TYY298 pKa = 11.73YY299 pKa = 11.05KK300 pKa = 10.88GADD303 pKa = 3.92CLSHH307 pKa = 6.36LTDD310 pKa = 3.51GAYY313 pKa = 9.36IEE315 pKa = 5.74LKK317 pKa = 10.21TNDD320 pKa = 5.09DD321 pKa = 3.35ISTATDD327 pKa = 3.21ISAVKK332 pKa = 10.39FEE334 pKa = 4.73MEE336 pKa = 3.88DD337 pKa = 2.91EE338 pKa = 4.8GYY340 pKa = 10.41EE341 pKa = 4.04DD342 pKa = 4.62HH343 pKa = 7.56SSDD346 pKa = 3.1SYY348 pKa = 11.32RR349 pKa = 11.84FEE351 pKa = 5.14IYY353 pKa = 10.83GKK355 pKa = 8.4TKK357 pKa = 10.84NIDD360 pKa = 3.38LTNGTFVFDD369 pKa = 3.89WNGTEE374 pKa = 3.78ITVSVDD380 pKa = 3.3TNIKK384 pKa = 10.67CEE386 pKa = 4.08INDD389 pKa = 3.33VYY391 pKa = 11.53YY392 pKa = 11.2YY393 pKa = 10.43GTDD396 pKa = 3.51CLNNLADD403 pKa = 4.05NLCIEE408 pKa = 5.34LKK410 pKa = 10.3TSDD413 pKa = 4.78DD414 pKa = 3.9VYY416 pKa = 11.29NYY418 pKa = 10.55DD419 pKa = 3.14GSAPITAVEE428 pKa = 4.72FEE430 pKa = 4.33TSDD433 pKa = 4.15DD434 pKa = 4.16CGTSYY439 pKa = 11.7
MM1 pKa = 7.61RR2 pKa = 11.84KK3 pKa = 8.96EE4 pKa = 4.15FKK6 pKa = 10.4KK7 pKa = 10.61KK8 pKa = 10.53AILGGSLSLSALLFACGGAGDD29 pKa = 4.14VASDD33 pKa = 3.58SPYY36 pKa = 9.86ATVTGTVTSASSTGSLSTTGIDD58 pKa = 2.89IGNVNLSFIAAVAIDD73 pKa = 3.94NGKK76 pKa = 9.49LVYY79 pKa = 9.44TADD82 pKa = 5.55DD83 pKa = 3.52IDD85 pKa = 4.3NNGNFNLKK93 pKa = 10.22LKK95 pKa = 10.51EE96 pKa = 4.08GLDD99 pKa = 3.54YY100 pKa = 11.57AFILFDD106 pKa = 3.35ISKK109 pKa = 9.9KK110 pKa = 9.99PKK112 pKa = 10.55LIVKK116 pKa = 9.75DD117 pKa = 3.89SNGNAIKK124 pKa = 10.59INGDD128 pKa = 3.12GSVNISLSDD137 pKa = 3.5SDD139 pKa = 4.84GDD141 pKa = 4.59GIPDD145 pKa = 3.67TASVNITGSVSLTKK159 pKa = 10.54DD160 pKa = 3.08SQLDD164 pKa = 3.84DD165 pKa = 4.41NDD167 pKa = 3.5NDD169 pKa = 4.29HH170 pKa = 6.7YY171 pKa = 11.23PDD173 pKa = 4.85GIEE176 pKa = 4.54NINAGGSVNPDD187 pKa = 2.98YY188 pKa = 11.6DD189 pKa = 3.83EE190 pKa = 6.35DD191 pKa = 5.49GDD193 pKa = 4.73GIFDD197 pKa = 5.24GIEE200 pKa = 4.13DD201 pKa = 4.34KK202 pKa = 11.5DD203 pKa = 3.72GDD205 pKa = 4.26GYY207 pKa = 10.71IDD209 pKa = 3.45GHH211 pKa = 6.31EE212 pKa = 4.67DD213 pKa = 3.45SNSNGIPDD221 pKa = 4.07VYY223 pKa = 11.1EE224 pKa = 5.72DD225 pKa = 4.62DD226 pKa = 5.92DD227 pKa = 6.15KK228 pKa = 12.02DD229 pKa = 3.8GLPNYY234 pKa = 10.38LDD236 pKa = 5.29DD237 pKa = 5.29SDD239 pKa = 5.27GDD241 pKa = 4.19GYY243 pKa = 9.96PDD245 pKa = 5.8HH246 pKa = 8.06IDD248 pKa = 3.64EE249 pKa = 5.8DD250 pKa = 4.79DD251 pKa = 3.76SDD253 pKa = 4.27GYY255 pKa = 10.66KK256 pKa = 10.28YY257 pKa = 10.79EE258 pKa = 4.16MKK260 pKa = 10.06GTVSNIDD267 pKa = 3.34TTTGTFVFTYY277 pKa = 10.58NGTDD281 pKa = 3.47YY282 pKa = 11.0NVSVTDD288 pKa = 3.53TTVCEE293 pKa = 4.46INDD296 pKa = 4.03TYY298 pKa = 11.73YY299 pKa = 11.05KK300 pKa = 10.88GADD303 pKa = 3.92CLSHH307 pKa = 6.36LTDD310 pKa = 3.51GAYY313 pKa = 9.36IEE315 pKa = 5.74LKK317 pKa = 10.21TNDD320 pKa = 5.09DD321 pKa = 3.35ISTATDD327 pKa = 3.21ISAVKK332 pKa = 10.39FEE334 pKa = 4.73MEE336 pKa = 3.88DD337 pKa = 2.91EE338 pKa = 4.8GYY340 pKa = 10.41EE341 pKa = 4.04DD342 pKa = 4.62HH343 pKa = 7.56SSDD346 pKa = 3.1SYY348 pKa = 11.32RR349 pKa = 11.84FEE351 pKa = 5.14IYY353 pKa = 10.83GKK355 pKa = 8.4TKK357 pKa = 10.84NIDD360 pKa = 3.38LTNGTFVFDD369 pKa = 3.89WNGTEE374 pKa = 3.78ITVSVDD380 pKa = 3.3TNIKK384 pKa = 10.67CEE386 pKa = 4.08INDD389 pKa = 3.33VYY391 pKa = 11.53YY392 pKa = 11.2YY393 pKa = 10.43GTDD396 pKa = 3.51CLNNLADD403 pKa = 4.05NLCIEE408 pKa = 5.34LKK410 pKa = 10.3TSDD413 pKa = 4.78DD414 pKa = 3.9VYY416 pKa = 11.29NYY418 pKa = 10.55DD419 pKa = 3.14GSAPITAVEE428 pKa = 4.72FEE430 pKa = 4.33TSDD433 pKa = 4.15DD434 pKa = 4.16CGTSYY439 pKa = 11.7
Molecular weight: 47.22 kDa
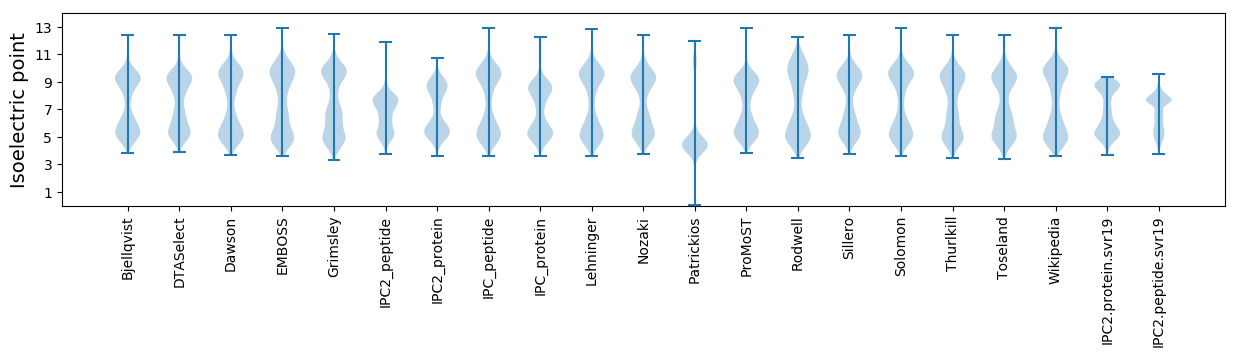
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|C0QQI0|C0QQI0_PERMH Sodium-driven multidrug efflux pump protein OS=Persephonella marina (strain DSM 14350 / EX-H1) OX=123214 GN=PERMA_1144 PE=4 SV=1
MM1 pKa = 8.07DD2 pKa = 3.3NTARR6 pKa = 11.84LDD8 pKa = 3.58YY9 pKa = 10.43PRR11 pKa = 11.84VKK13 pKa = 10.79GGISPAAPGTPKK25 pKa = 10.59GALPSLPPILRR36 pKa = 11.84MTRR39 pKa = 11.84KK40 pKa = 9.71QSMPGYY46 pKa = 8.93SKK48 pKa = 11.05ASRR51 pKa = 11.84GLSVLPRR58 pKa = 11.84VVGILTDD65 pKa = 3.62TTISPGLSSRR75 pKa = 11.84QRR77 pKa = 11.84GDD79 pKa = 2.16RR80 pKa = 11.84WTIHH84 pKa = 6.78AGRR87 pKa = 11.84NLPDD91 pKa = 3.14KK92 pKa = 10.04EE93 pKa = 4.13FRR95 pKa = 11.84YY96 pKa = 10.06HH97 pKa = 5.39RR98 pKa = 11.84TVIVTAAVYY107 pKa = 9.81PGFGSRR113 pKa = 11.84LAPLPLTYY121 pKa = 10.47GHH123 pKa = 7.22WAGFTPHH130 pKa = 6.41TSSFEE135 pKa = 3.91FAQCCVFGKK144 pKa = 10.15QSPPPGFCDD153 pKa = 4.44PLPHH157 pKa = 7.2LNGTACGHH165 pKa = 6.22PLSRR169 pKa = 11.84SYY171 pKa = 11.23GVNLPSSLRR180 pKa = 11.84RR181 pKa = 11.84VTPTALGYY189 pKa = 10.69
MM1 pKa = 8.07DD2 pKa = 3.3NTARR6 pKa = 11.84LDD8 pKa = 3.58YY9 pKa = 10.43PRR11 pKa = 11.84VKK13 pKa = 10.79GGISPAAPGTPKK25 pKa = 10.59GALPSLPPILRR36 pKa = 11.84MTRR39 pKa = 11.84KK40 pKa = 9.71QSMPGYY46 pKa = 8.93SKK48 pKa = 11.05ASRR51 pKa = 11.84GLSVLPRR58 pKa = 11.84VVGILTDD65 pKa = 3.62TTISPGLSSRR75 pKa = 11.84QRR77 pKa = 11.84GDD79 pKa = 2.16RR80 pKa = 11.84WTIHH84 pKa = 6.78AGRR87 pKa = 11.84NLPDD91 pKa = 3.14KK92 pKa = 10.04EE93 pKa = 4.13FRR95 pKa = 11.84YY96 pKa = 10.06HH97 pKa = 5.39RR98 pKa = 11.84TVIVTAAVYY107 pKa = 9.81PGFGSRR113 pKa = 11.84LAPLPLTYY121 pKa = 10.47GHH123 pKa = 7.22WAGFTPHH130 pKa = 6.41TSSFEE135 pKa = 3.91FAQCCVFGKK144 pKa = 10.15QSPPPGFCDD153 pKa = 4.44PLPHH157 pKa = 7.2LNGTACGHH165 pKa = 6.22PLSRR169 pKa = 11.84SYY171 pKa = 11.23GVNLPSSLRR180 pKa = 11.84RR181 pKa = 11.84VTPTALGYY189 pKa = 10.69
Molecular weight: 20.38 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
622608 |
37 |
5809 |
304.0 |
34.63 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.536 ± 0.062 | 0.742 ± 0.023 |
5.699 ± 0.044 | 8.178 ± 0.077 |
4.992 ± 0.057 | 6.49 ± 0.056 |
1.507 ± 0.025 | 9.36 ± 0.053 |
9.185 ± 0.07 | 9.283 ± 0.054 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.999 ± 0.024 | 4.241 ± 0.061 |
3.671 ± 0.039 | 2.328 ± 0.03 |
4.664 ± 0.044 | 5.676 ± 0.071 |
4.562 ± 0.042 | 6.839 ± 0.044 |
0.804 ± 0.021 | 4.245 ± 0.037 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
