
Xanthomonas citri phage CP2
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Podoviridae; unclassified Podoviridae
Average proteome isoelectric point is 6.28
Get precalculated fractions of proteins
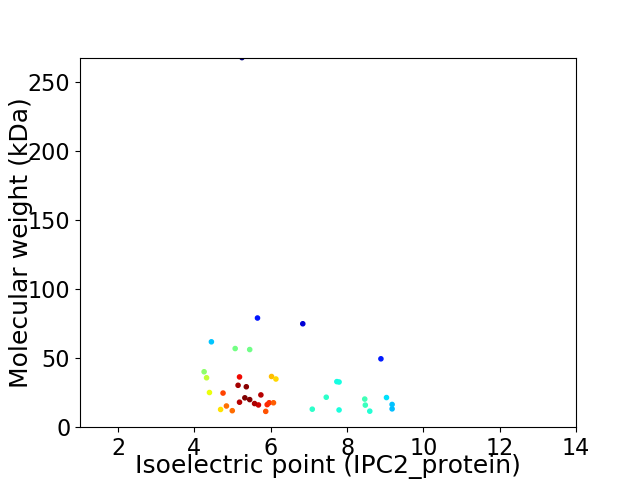
Virtual 2D-PAGE plot for 40 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
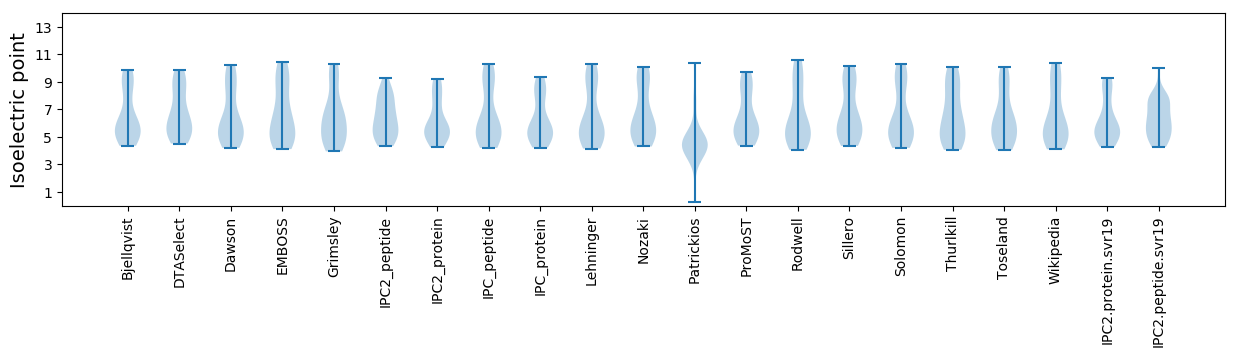
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|K7ZLE9|K7ZLE9_9CAUD Uncharacterized protein OS=Xanthomonas citri phage CP2 OX=1188795 PE=4 SV=1
MM1 pKa = 7.03TVSANSRR8 pKa = 11.84RR9 pKa = 11.84RR10 pKa = 11.84EE11 pKa = 3.87YY12 pKa = 10.53QGNGVTQVFNGPMAYY27 pKa = 10.1ARR29 pKa = 11.84AHH31 pKa = 5.77VYY33 pKa = 10.96AFLVKK38 pKa = 9.93GKK40 pKa = 8.85TMTLVPNASFDD51 pKa = 3.94VEE53 pKa = 4.15KK54 pKa = 10.83LGYY57 pKa = 10.12EE58 pKa = 3.77SGTRR62 pKa = 11.84VTMHH66 pKa = 6.87TPPAADD72 pKa = 3.58EE73 pKa = 4.09MLLLLRR79 pKa = 11.84TMPYY83 pKa = 8.38TQEE86 pKa = 3.69TDD88 pKa = 2.79ITNQGAFHH96 pKa = 7.35AEE98 pKa = 4.19TIEE101 pKa = 4.04KK102 pKa = 10.63GFDD105 pKa = 3.36ALEE108 pKa = 3.93MQIQQLVDD116 pKa = 3.25GTIQLVFEE124 pKa = 5.26DD125 pKa = 6.01GEE127 pKa = 4.68FVWDD131 pKa = 3.88AKK133 pKa = 10.24GSRR136 pKa = 11.84IIRR139 pKa = 11.84VGDD142 pKa = 3.35PTGDD146 pKa = 3.15QDD148 pKa = 4.64AVNLRR153 pKa = 11.84TLYY156 pKa = 10.89LVIEE160 pKa = 4.36QIQNGGGSVGVSPKK174 pKa = 9.62YY175 pKa = 10.11WEE177 pKa = 3.86ITGDD181 pKa = 4.02GEE183 pKa = 4.54DD184 pKa = 4.21TDD186 pKa = 5.37FEE188 pKa = 5.0LPGSDD193 pKa = 3.69VSDD196 pKa = 3.54LLFYY200 pKa = 11.12DD201 pKa = 4.05VVVEE205 pKa = 4.24GLSKK209 pKa = 10.69EE210 pKa = 4.37PYY212 pKa = 9.9DD213 pKa = 3.92EE214 pKa = 4.17YY215 pKa = 11.18TIVAASDD222 pKa = 3.6GSDD225 pKa = 3.29FAVRR229 pKa = 11.84FPAPLPNGAEE239 pKa = 4.0GFVILRR245 pKa = 11.84GYY247 pKa = 10.62AKK249 pKa = 10.0PFTGNTPVQTLRR261 pKa = 11.84IPIIDD266 pKa = 3.73VDD268 pKa = 3.65ATTYY272 pKa = 10.31TVDD275 pKa = 3.34KK276 pKa = 10.54SSEE279 pKa = 3.96FALLRR284 pKa = 11.84TTNVNPCLLTLRR296 pKa = 11.84ANQGDD301 pKa = 4.62LFDD304 pKa = 4.91MGDD307 pKa = 3.0GSYY310 pKa = 11.07FSVCQRR316 pKa = 11.84GDD318 pKa = 3.12GQARR322 pKa = 11.84IVADD326 pKa = 3.42TGVTLVLPEE335 pKa = 4.08GFVPYY340 pKa = 9.63TRR342 pKa = 11.84APGSIISAVCEE353 pKa = 4.01YY354 pKa = 11.34AEE356 pKa = 4.35GDD358 pKa = 3.61TWVLSGDD365 pKa = 4.04LAQGG369 pKa = 3.38
MM1 pKa = 7.03TVSANSRR8 pKa = 11.84RR9 pKa = 11.84RR10 pKa = 11.84EE11 pKa = 3.87YY12 pKa = 10.53QGNGVTQVFNGPMAYY27 pKa = 10.1ARR29 pKa = 11.84AHH31 pKa = 5.77VYY33 pKa = 10.96AFLVKK38 pKa = 9.93GKK40 pKa = 8.85TMTLVPNASFDD51 pKa = 3.94VEE53 pKa = 4.15KK54 pKa = 10.83LGYY57 pKa = 10.12EE58 pKa = 3.77SGTRR62 pKa = 11.84VTMHH66 pKa = 6.87TPPAADD72 pKa = 3.58EE73 pKa = 4.09MLLLLRR79 pKa = 11.84TMPYY83 pKa = 8.38TQEE86 pKa = 3.69TDD88 pKa = 2.79ITNQGAFHH96 pKa = 7.35AEE98 pKa = 4.19TIEE101 pKa = 4.04KK102 pKa = 10.63GFDD105 pKa = 3.36ALEE108 pKa = 3.93MQIQQLVDD116 pKa = 3.25GTIQLVFEE124 pKa = 5.26DD125 pKa = 6.01GEE127 pKa = 4.68FVWDD131 pKa = 3.88AKK133 pKa = 10.24GSRR136 pKa = 11.84IIRR139 pKa = 11.84VGDD142 pKa = 3.35PTGDD146 pKa = 3.15QDD148 pKa = 4.64AVNLRR153 pKa = 11.84TLYY156 pKa = 10.89LVIEE160 pKa = 4.36QIQNGGGSVGVSPKK174 pKa = 9.62YY175 pKa = 10.11WEE177 pKa = 3.86ITGDD181 pKa = 4.02GEE183 pKa = 4.54DD184 pKa = 4.21TDD186 pKa = 5.37FEE188 pKa = 5.0LPGSDD193 pKa = 3.69VSDD196 pKa = 3.54LLFYY200 pKa = 11.12DD201 pKa = 4.05VVVEE205 pKa = 4.24GLSKK209 pKa = 10.69EE210 pKa = 4.37PYY212 pKa = 9.9DD213 pKa = 3.92EE214 pKa = 4.17YY215 pKa = 11.18TIVAASDD222 pKa = 3.6GSDD225 pKa = 3.29FAVRR229 pKa = 11.84FPAPLPNGAEE239 pKa = 4.0GFVILRR245 pKa = 11.84GYY247 pKa = 10.62AKK249 pKa = 10.0PFTGNTPVQTLRR261 pKa = 11.84IPIIDD266 pKa = 3.73VDD268 pKa = 3.65ATTYY272 pKa = 10.31TVDD275 pKa = 3.34KK276 pKa = 10.54SSEE279 pKa = 3.96FALLRR284 pKa = 11.84TTNVNPCLLTLRR296 pKa = 11.84ANQGDD301 pKa = 4.62LFDD304 pKa = 4.91MGDD307 pKa = 3.0GSYY310 pKa = 11.07FSVCQRR316 pKa = 11.84GDD318 pKa = 3.12GQARR322 pKa = 11.84IVADD326 pKa = 3.42TGVTLVLPEE335 pKa = 4.08GFVPYY340 pKa = 9.63TRR342 pKa = 11.84APGSIISAVCEE353 pKa = 4.01YY354 pKa = 11.34AEE356 pKa = 4.35GDD358 pKa = 3.61TWVLSGDD365 pKa = 4.04LAQGG369 pKa = 3.38
Molecular weight: 40.13 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|K7ZJS9|K7ZJS9_9CAUD Uncharacterized protein OS=Xanthomonas citri phage CP2 OX=1188795 PE=4 SV=1
MM1 pKa = 7.56GARR4 pKa = 11.84TPSPPYY10 pKa = 8.56PRR12 pKa = 11.84MMKK15 pKa = 9.84RR16 pKa = 11.84VRR18 pKa = 11.84VAEE21 pKa = 4.28SGCWLYY27 pKa = 10.73TGSKK31 pKa = 10.41DD32 pKa = 3.14GGGYY36 pKa = 8.34GTVSIRR42 pKa = 11.84RR43 pKa = 11.84GAAPGKK49 pKa = 9.25AHH51 pKa = 7.28RR52 pKa = 11.84ISYY55 pKa = 7.13EE56 pKa = 3.83HH57 pKa = 6.94HH58 pKa = 6.37VGAIPVGAMVCHH70 pKa = 6.52RR71 pKa = 11.84CDD73 pKa = 3.09TPACVNPDD81 pKa = 3.16HH82 pKa = 7.01LFIGTQGDD90 pKa = 3.59NMQDD94 pKa = 3.49CASKK98 pKa = 10.94GRR100 pKa = 11.84ISPRR104 pKa = 11.84SILNLQPGAPGHH116 pKa = 6.6HH117 pKa = 6.19GAAPRR122 pKa = 11.84KK123 pKa = 8.56QAA125 pKa = 3.65
MM1 pKa = 7.56GARR4 pKa = 11.84TPSPPYY10 pKa = 8.56PRR12 pKa = 11.84MMKK15 pKa = 9.84RR16 pKa = 11.84VRR18 pKa = 11.84VAEE21 pKa = 4.28SGCWLYY27 pKa = 10.73TGSKK31 pKa = 10.41DD32 pKa = 3.14GGGYY36 pKa = 8.34GTVSIRR42 pKa = 11.84RR43 pKa = 11.84GAAPGKK49 pKa = 9.25AHH51 pKa = 7.28RR52 pKa = 11.84ISYY55 pKa = 7.13EE56 pKa = 3.83HH57 pKa = 6.94HH58 pKa = 6.37VGAIPVGAMVCHH70 pKa = 6.52RR71 pKa = 11.84CDD73 pKa = 3.09TPACVNPDD81 pKa = 3.16HH82 pKa = 7.01LFIGTQGDD90 pKa = 3.59NMQDD94 pKa = 3.49CASKK98 pKa = 10.94GRR100 pKa = 11.84ISPRR104 pKa = 11.84SILNLQPGAPGHH116 pKa = 6.6HH117 pKa = 6.19GAAPRR122 pKa = 11.84KK123 pKa = 8.56QAA125 pKa = 3.65
Molecular weight: 13.27 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
12684 |
104 |
2478 |
317.1 |
34.26 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.679 ± 0.819 | 0.922 ± 0.295 |
6.457 ± 0.335 | 5.637 ± 0.326 |
2.996 ± 0.202 | 8.538 ± 0.369 |
1.451 ± 0.146 | 4.06 ± 0.276 |
3.784 ± 0.269 | 7.411 ± 0.397 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.294 ± 0.127 | 3.319 ± 0.233 |
5.503 ± 0.647 | 4.857 ± 0.287 |
6.875 ± 0.38 | 5.361 ± 0.335 |
5.905 ± 0.261 | 7.017 ± 0.301 |
1.632 ± 0.205 | 2.302 ± 0.156 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
