
Staphylococcus microti
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Bacillales; Staphylococcaceae; Staphylococcus
Average proteome isoelectric point is 6.27
Get precalculated fractions of proteins
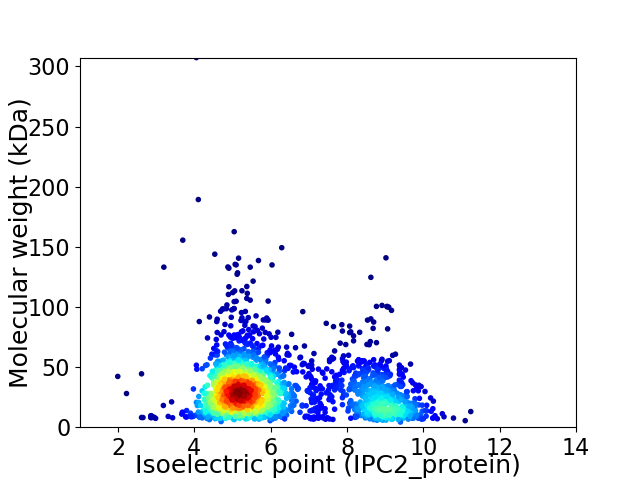
Virtual 2D-PAGE plot for 2166 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
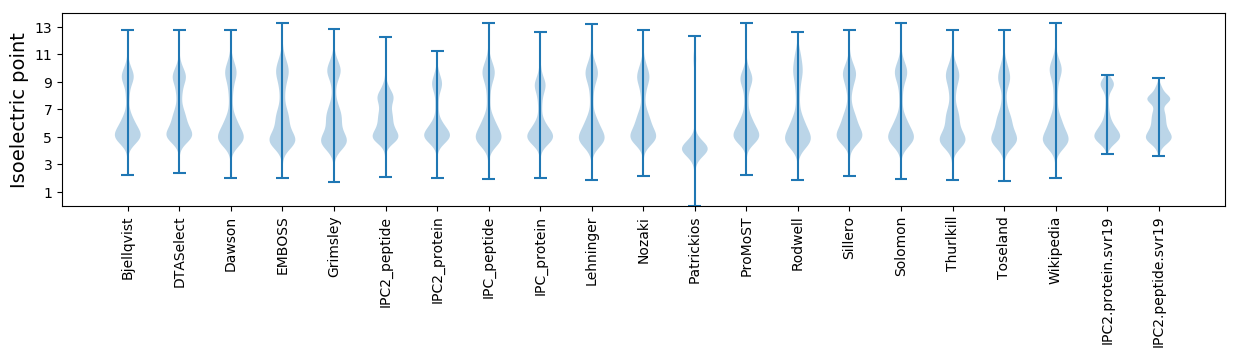
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0D6XS18|A0A0D6XS18_9STAP Zinc metalloprotease OS=Staphylococcus microti OX=569857 GN=NCTC13832_00988 PE=3 SV=1
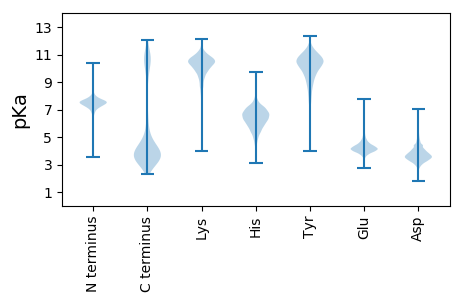
MM1 pKa = 7.67KK2 pKa = 10.13KK3 pKa = 10.15QSFLVGVVASTLLLTGCNFLTPNNQNNNQSTNGANNQSSNQNNQQNTNGQGSYY56 pKa = 10.04IYY58 pKa = 10.04DD59 pKa = 3.39QNTRR63 pKa = 11.84EE64 pKa = 4.14YY65 pKa = 10.51YY66 pKa = 9.16AQIWLTVRR74 pKa = 11.84DD75 pKa = 4.02NIEE78 pKa = 3.92TFNQGDD84 pKa = 3.62VSYY87 pKa = 10.25TPVDD91 pKa = 3.49VAGTLVNPYY100 pKa = 9.99NADD103 pKa = 3.1ATATYY108 pKa = 10.32PEE110 pKa = 4.37GTITLSASPTAAGSITFKK128 pKa = 10.74DD129 pKa = 3.54NHH131 pKa = 6.98DD132 pKa = 3.79GTITVYY138 pKa = 10.18NAPGHH143 pKa = 5.52FHH145 pKa = 7.65DD146 pKa = 5.94DD147 pKa = 2.76RR148 pKa = 11.84WFEE151 pKa = 4.48DD152 pKa = 3.73DD153 pKa = 3.69FSLRR157 pKa = 11.84EE158 pKa = 3.92SQKK161 pKa = 10.86IIDD164 pKa = 3.83NGYY167 pKa = 7.73TLTIKK172 pKa = 10.45NASQSDD178 pKa = 3.6INYY181 pKa = 7.33VAQYY185 pKa = 8.27ITQSTPQPSPSEE197 pKa = 4.06AYY199 pKa = 9.79EE200 pKa = 4.84GIDD203 pKa = 3.19SGRR206 pKa = 11.84EE207 pKa = 3.74EE208 pKa = 4.37SSSSEE213 pKa = 3.81DD214 pKa = 3.43SEE216 pKa = 4.3ITVTRR221 pKa = 11.84DD222 pKa = 3.09NVIDD226 pKa = 3.92LVEE229 pKa = 4.83AYY231 pKa = 9.86EE232 pKa = 4.95GDD234 pKa = 4.0FLDD237 pKa = 3.78TDD239 pKa = 4.01TYY241 pKa = 10.25TFKK244 pKa = 10.9EE245 pKa = 4.4PEE247 pKa = 4.1KK248 pKa = 10.37MSDD251 pKa = 3.64GSWGFSILDD260 pKa = 3.62KK261 pKa = 10.6EE262 pKa = 4.61TGEE265 pKa = 4.19LAGSYY270 pKa = 10.14IIDD273 pKa = 4.07PDD275 pKa = 3.98GTVTKK280 pKa = 10.37YY281 pKa = 10.63DD282 pKa = 3.31EE283 pKa = 4.96HH284 pKa = 7.2GNPEE288 pKa = 3.79
MM1 pKa = 7.67KK2 pKa = 10.13KK3 pKa = 10.15QSFLVGVVASTLLLTGCNFLTPNNQNNNQSTNGANNQSSNQNNQQNTNGQGSYY56 pKa = 10.04IYY58 pKa = 10.04DD59 pKa = 3.39QNTRR63 pKa = 11.84EE64 pKa = 4.14YY65 pKa = 10.51YY66 pKa = 9.16AQIWLTVRR74 pKa = 11.84DD75 pKa = 4.02NIEE78 pKa = 3.92TFNQGDD84 pKa = 3.62VSYY87 pKa = 10.25TPVDD91 pKa = 3.49VAGTLVNPYY100 pKa = 9.99NADD103 pKa = 3.1ATATYY108 pKa = 10.32PEE110 pKa = 4.37GTITLSASPTAAGSITFKK128 pKa = 10.74DD129 pKa = 3.54NHH131 pKa = 6.98DD132 pKa = 3.79GTITVYY138 pKa = 10.18NAPGHH143 pKa = 5.52FHH145 pKa = 7.65DD146 pKa = 5.94DD147 pKa = 2.76RR148 pKa = 11.84WFEE151 pKa = 4.48DD152 pKa = 3.73DD153 pKa = 3.69FSLRR157 pKa = 11.84EE158 pKa = 3.92SQKK161 pKa = 10.86IIDD164 pKa = 3.83NGYY167 pKa = 7.73TLTIKK172 pKa = 10.45NASQSDD178 pKa = 3.6INYY181 pKa = 7.33VAQYY185 pKa = 8.27ITQSTPQPSPSEE197 pKa = 4.06AYY199 pKa = 9.79EE200 pKa = 4.84GIDD203 pKa = 3.19SGRR206 pKa = 11.84EE207 pKa = 3.74EE208 pKa = 4.37SSSSEE213 pKa = 3.81DD214 pKa = 3.43SEE216 pKa = 4.3ITVTRR221 pKa = 11.84DD222 pKa = 3.09NVIDD226 pKa = 3.92LVEE229 pKa = 4.83AYY231 pKa = 9.86EE232 pKa = 4.95GDD234 pKa = 4.0FLDD237 pKa = 3.78TDD239 pKa = 4.01TYY241 pKa = 10.25TFKK244 pKa = 10.9EE245 pKa = 4.4PEE247 pKa = 4.1KK248 pKa = 10.37MSDD251 pKa = 3.64GSWGFSILDD260 pKa = 3.62KK261 pKa = 10.6EE262 pKa = 4.61TGEE265 pKa = 4.19LAGSYY270 pKa = 10.14IIDD273 pKa = 4.07PDD275 pKa = 3.98GTVTKK280 pKa = 10.37YY281 pKa = 10.63DD282 pKa = 3.31EE283 pKa = 4.96HH284 pKa = 7.2GNPEE288 pKa = 3.79
Molecular weight: 31.79 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0D6XN60|A0A0D6XN60_9STAP ATP-dependent zinc metalloprotease FtsH OS=Staphylococcus microti OX=569857 GN=ftsH PE=3 SV=1
MM1 pKa = 7.38KK2 pKa = 9.94VQRR5 pKa = 11.84IVIQQKK11 pKa = 8.25HH12 pKa = 4.4QKK14 pKa = 10.12ANQLKK19 pKa = 9.59MMRR22 pKa = 11.84QHH24 pKa = 5.78QLNRR28 pKa = 11.84TVNRR32 pKa = 11.84LLNQKK37 pKa = 9.25VRR39 pKa = 11.84VRR41 pKa = 11.84LNQYY45 pKa = 9.73LFQRR49 pKa = 11.84ASKK52 pKa = 10.02LQHH55 pKa = 5.18QKK57 pKa = 10.05VRR59 pKa = 11.84VHH61 pKa = 5.87QLQKK65 pKa = 10.52VKK67 pKa = 10.47KK68 pKa = 7.57HH69 pKa = 4.4QKK71 pKa = 8.77VRR73 pKa = 11.84KK74 pKa = 8.19QRR76 pKa = 11.84LRR78 pKa = 11.84IQKK81 pKa = 7.7QHH83 pKa = 5.19QRR85 pKa = 11.84AKK87 pKa = 9.23RR88 pKa = 11.84QVTVTHH94 pKa = 5.86NHH96 pKa = 4.44QMFPRR101 pKa = 11.84PQTAA105 pKa = 2.73
MM1 pKa = 7.38KK2 pKa = 9.94VQRR5 pKa = 11.84IVIQQKK11 pKa = 8.25HH12 pKa = 4.4QKK14 pKa = 10.12ANQLKK19 pKa = 9.59MMRR22 pKa = 11.84QHH24 pKa = 5.78QLNRR28 pKa = 11.84TVNRR32 pKa = 11.84LLNQKK37 pKa = 9.25VRR39 pKa = 11.84VRR41 pKa = 11.84LNQYY45 pKa = 9.73LFQRR49 pKa = 11.84ASKK52 pKa = 10.02LQHH55 pKa = 5.18QKK57 pKa = 10.05VRR59 pKa = 11.84VHH61 pKa = 5.87QLQKK65 pKa = 10.52VKK67 pKa = 10.47KK68 pKa = 7.57HH69 pKa = 4.4QKK71 pKa = 8.77VRR73 pKa = 11.84KK74 pKa = 8.19QRR76 pKa = 11.84LRR78 pKa = 11.84IQKK81 pKa = 7.7QHH83 pKa = 5.19QRR85 pKa = 11.84AKK87 pKa = 9.23RR88 pKa = 11.84QVTVTHH94 pKa = 5.86NHH96 pKa = 4.44QMFPRR101 pKa = 11.84PQTAA105 pKa = 2.73
Molecular weight: 13.0 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
640098 |
37 |
2944 |
295.5 |
33.22 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.378 ± 0.088 | 0.625 ± 0.016 |
5.982 ± 0.08 | 6.643 ± 0.073 |
4.312 ± 0.053 | 6.329 ± 0.052 |
2.494 ± 0.029 | 7.724 ± 0.061 |
6.439 ± 0.06 | 9.402 ± 0.063 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.819 ± 0.03 | 4.417 ± 0.036 |
3.486 ± 0.038 | 4.503 ± 0.044 |
4.015 ± 0.041 | 5.523 ± 0.073 |
6.039 ± 0.041 | 7.225 ± 0.042 |
0.761 ± 0.018 | 3.886 ± 0.042 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
