
Tenacibaculum sp. MAR_2009_124
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Tenacibaculum; unclassified Tenacibaculum
Average proteome isoelectric point is 6.86
Get precalculated fractions of proteins
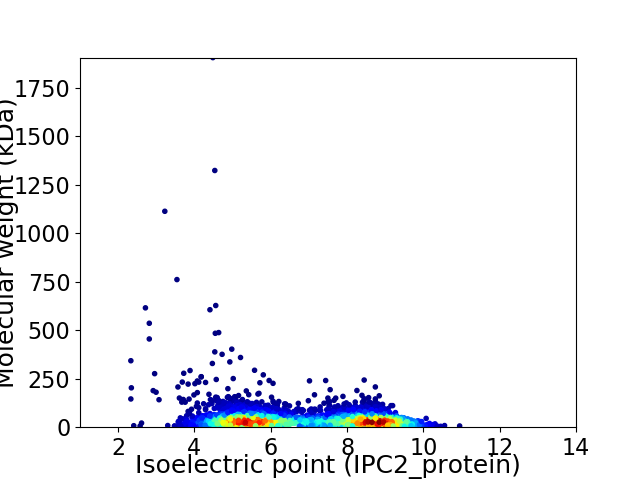
Virtual 2D-PAGE plot for 4480 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1H4JS34|A0A1H4JS34_9FLAO Uncharacterized protein OS=Tenacibaculum sp. MAR_2009_124 OX=1250059 GN=SAMN04489761_0998 PE=4 SV=1
MM1 pKa = 6.13VTFVFVLGFIACEE14 pKa = 4.1DD15 pKa = 3.64NDD17 pKa = 3.74NLEE20 pKa = 4.36FTVQAPQDD28 pKa = 3.73SVAFTNTLLNEE39 pKa = 4.36YY40 pKa = 9.59IITPQTSGNIAEE52 pKa = 4.08RR53 pKa = 11.84FVWNSVVFIGTATEE67 pKa = 3.92VTYY70 pKa = 10.73DD71 pKa = 3.69LQVSITQDD79 pKa = 3.24FASFEE84 pKa = 4.6SLGTTNDD91 pKa = 2.66THH93 pKa = 6.91MSVTVDD99 pKa = 3.26KK100 pKa = 10.98LLALAEE106 pKa = 4.25TAGLDD111 pKa = 3.94NDD113 pKa = 4.17PTTDD117 pKa = 3.87DD118 pKa = 4.36KK119 pKa = 11.91PNMGLVYY126 pKa = 9.79FRR128 pKa = 11.84VRR130 pKa = 11.84AFVGNGGTDD139 pKa = 3.39APEE142 pKa = 4.71SISDD146 pKa = 3.41IMVMNITLPEE156 pKa = 3.86ITQGSGIEE164 pKa = 3.85ISSWGIVGSGYY175 pKa = 10.6NDD177 pKa = 3.48WGNAGPDD184 pKa = 3.23APFYY188 pKa = 7.42TTNTANVLVAYY199 pKa = 7.51VTLLDD204 pKa = 3.99GEE206 pKa = 4.41IKK208 pKa = 10.58LRR210 pKa = 11.84EE211 pKa = 4.04NNEE214 pKa = 3.65WTNNLGDD221 pKa = 4.82DD222 pKa = 4.27GADD225 pKa = 3.3GTLEE229 pKa = 3.89VGGANIVSSAGTYY242 pKa = 10.29KK243 pKa = 9.28ITLDD247 pKa = 4.27LNTNTYY253 pKa = 9.63TIEE256 pKa = 4.16SYY258 pKa = 10.23SWGIIGSGFNDD269 pKa = 3.86WGNDD273 pKa = 3.52GPDD276 pKa = 3.19AEE278 pKa = 4.3FHH280 pKa = 6.48YY281 pKa = 10.6DD282 pKa = 3.36YY283 pKa = 8.57TTDD286 pKa = 3.31TFKK289 pKa = 11.38VGVKK293 pKa = 10.51LLDD296 pKa = 3.65GEE298 pKa = 4.58IKK300 pKa = 10.42FRR302 pKa = 11.84LNNNWTTNYY311 pKa = 10.6GGTDD315 pKa = 3.31GNLAAGGDD323 pKa = 4.3NIVSTAGFYY332 pKa = 10.51QVTIDD337 pKa = 4.31FNNNTYY343 pKa = 9.92TIEE346 pKa = 4.47ANDD349 pKa = 3.11VWGIIGSGYY358 pKa = 10.11NDD360 pKa = 3.75WGNNGPDD367 pKa = 3.61FNFTQVNPGIYY378 pKa = 9.34IANNVTLLDD387 pKa = 3.84GEE389 pKa = 4.39IKK391 pKa = 10.44FRR393 pKa = 11.84LNEE396 pKa = 3.87DD397 pKa = 2.81WTTNFGDD404 pKa = 4.58DD405 pKa = 4.27GNDD408 pKa = 3.38GTLEE412 pKa = 4.12AEE414 pKa = 4.67GANIPSTAGKK424 pKa = 9.95FRR426 pKa = 11.84ITMDD430 pKa = 4.09LSDD433 pKa = 4.59NSNPTYY439 pKa = 10.26TISSLL444 pKa = 3.53
MM1 pKa = 6.13VTFVFVLGFIACEE14 pKa = 4.1DD15 pKa = 3.64NDD17 pKa = 3.74NLEE20 pKa = 4.36FTVQAPQDD28 pKa = 3.73SVAFTNTLLNEE39 pKa = 4.36YY40 pKa = 9.59IITPQTSGNIAEE52 pKa = 4.08RR53 pKa = 11.84FVWNSVVFIGTATEE67 pKa = 3.92VTYY70 pKa = 10.73DD71 pKa = 3.69LQVSITQDD79 pKa = 3.24FASFEE84 pKa = 4.6SLGTTNDD91 pKa = 2.66THH93 pKa = 6.91MSVTVDD99 pKa = 3.26KK100 pKa = 10.98LLALAEE106 pKa = 4.25TAGLDD111 pKa = 3.94NDD113 pKa = 4.17PTTDD117 pKa = 3.87DD118 pKa = 4.36KK119 pKa = 11.91PNMGLVYY126 pKa = 9.79FRR128 pKa = 11.84VRR130 pKa = 11.84AFVGNGGTDD139 pKa = 3.39APEE142 pKa = 4.71SISDD146 pKa = 3.41IMVMNITLPEE156 pKa = 3.86ITQGSGIEE164 pKa = 3.85ISSWGIVGSGYY175 pKa = 10.6NDD177 pKa = 3.48WGNAGPDD184 pKa = 3.23APFYY188 pKa = 7.42TTNTANVLVAYY199 pKa = 7.51VTLLDD204 pKa = 3.99GEE206 pKa = 4.41IKK208 pKa = 10.58LRR210 pKa = 11.84EE211 pKa = 4.04NNEE214 pKa = 3.65WTNNLGDD221 pKa = 4.82DD222 pKa = 4.27GADD225 pKa = 3.3GTLEE229 pKa = 3.89VGGANIVSSAGTYY242 pKa = 10.29KK243 pKa = 9.28ITLDD247 pKa = 4.27LNTNTYY253 pKa = 9.63TIEE256 pKa = 4.16SYY258 pKa = 10.23SWGIIGSGFNDD269 pKa = 3.86WGNDD273 pKa = 3.52GPDD276 pKa = 3.19AEE278 pKa = 4.3FHH280 pKa = 6.48YY281 pKa = 10.6DD282 pKa = 3.36YY283 pKa = 8.57TTDD286 pKa = 3.31TFKK289 pKa = 11.38VGVKK293 pKa = 10.51LLDD296 pKa = 3.65GEE298 pKa = 4.58IKK300 pKa = 10.42FRR302 pKa = 11.84LNNNWTTNYY311 pKa = 10.6GGTDD315 pKa = 3.31GNLAAGGDD323 pKa = 4.3NIVSTAGFYY332 pKa = 10.51QVTIDD337 pKa = 4.31FNNNTYY343 pKa = 9.92TIEE346 pKa = 4.47ANDD349 pKa = 3.11VWGIIGSGYY358 pKa = 10.11NDD360 pKa = 3.75WGNNGPDD367 pKa = 3.61FNFTQVNPGIYY378 pKa = 9.34IANNVTLLDD387 pKa = 3.84GEE389 pKa = 4.39IKK391 pKa = 10.44FRR393 pKa = 11.84LNEE396 pKa = 3.87DD397 pKa = 2.81WTTNFGDD404 pKa = 4.58DD405 pKa = 4.27GNDD408 pKa = 3.38GTLEE412 pKa = 4.12AEE414 pKa = 4.67GANIPSTAGKK424 pKa = 9.95FRR426 pKa = 11.84ITMDD430 pKa = 4.09LSDD433 pKa = 4.59NSNPTYY439 pKa = 10.26TISSLL444 pKa = 3.53
Molecular weight: 48.1 kDa
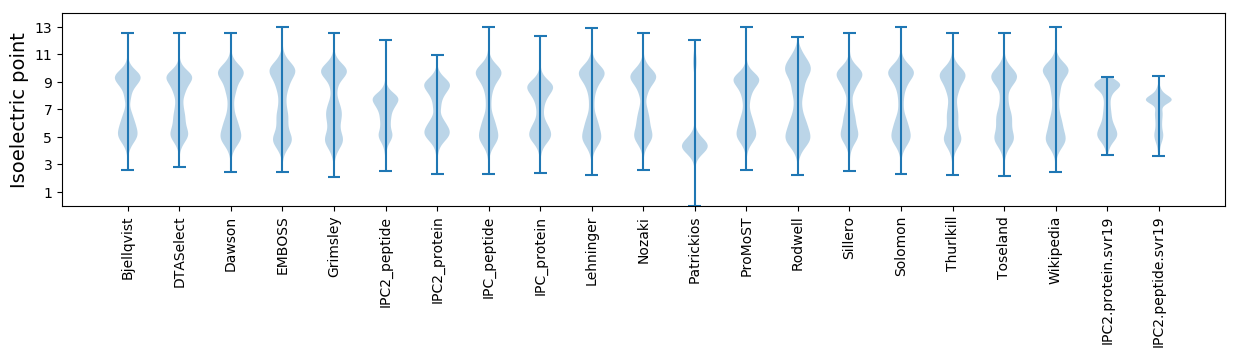
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1H4IZP7|A0A1H4IZP7_9FLAO Phosphoribosylaminoimidazole-succinocarboxamide synthase OS=Tenacibaculum sp. MAR_2009_124 OX=1250059 GN=purC PE=3 SV=1
MM1 pKa = 7.74SKK3 pKa = 8.95RR4 pKa = 11.84TYY6 pKa = 10.24QPSKK10 pKa = 9.01RR11 pKa = 11.84KK12 pKa = 9.47RR13 pKa = 11.84RR14 pKa = 11.84NKK16 pKa = 9.49HH17 pKa = 3.94GFRR20 pKa = 11.84EE21 pKa = 4.27RR22 pKa = 11.84MASANGRR29 pKa = 11.84KK30 pKa = 9.04VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 9.73GRR40 pKa = 11.84KK41 pKa = 8.36RR42 pKa = 11.84LTVSSQARR50 pKa = 11.84PKK52 pKa = 10.34KK53 pKa = 10.45
MM1 pKa = 7.74SKK3 pKa = 8.95RR4 pKa = 11.84TYY6 pKa = 10.24QPSKK10 pKa = 9.01RR11 pKa = 11.84KK12 pKa = 9.47RR13 pKa = 11.84RR14 pKa = 11.84NKK16 pKa = 9.49HH17 pKa = 3.94GFRR20 pKa = 11.84EE21 pKa = 4.27RR22 pKa = 11.84MASANGRR29 pKa = 11.84KK30 pKa = 9.04VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 9.73GRR40 pKa = 11.84KK41 pKa = 8.36RR42 pKa = 11.84LTVSSQARR50 pKa = 11.84PKK52 pKa = 10.34KK53 pKa = 10.45
Molecular weight: 6.31 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1587372 |
28 |
17153 |
354.3 |
40.04 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.424 ± 0.043 | 0.8 ± 0.016 |
5.638 ± 0.101 | 6.697 ± 0.06 |
5.147 ± 0.048 | 6.319 ± 0.057 |
1.725 ± 0.02 | 8.055 ± 0.045 |
8.145 ± 0.102 | 9.107 ± 0.069 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.889 ± 0.022 | 6.847 ± 0.044 |
3.213 ± 0.03 | 3.199 ± 0.026 |
3.26 ± 0.037 | 7.101 ± 0.045 |
6.061 ± 0.098 | 6.188 ± 0.04 |
1.015 ± 0.014 | 4.17 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
