
Tomato mottle mosaic virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Alsuviricetes; Martellivirales; Virgaviridae; Tobamovirus
Average proteome isoelectric point is 5.95
Get precalculated fractions of proteins
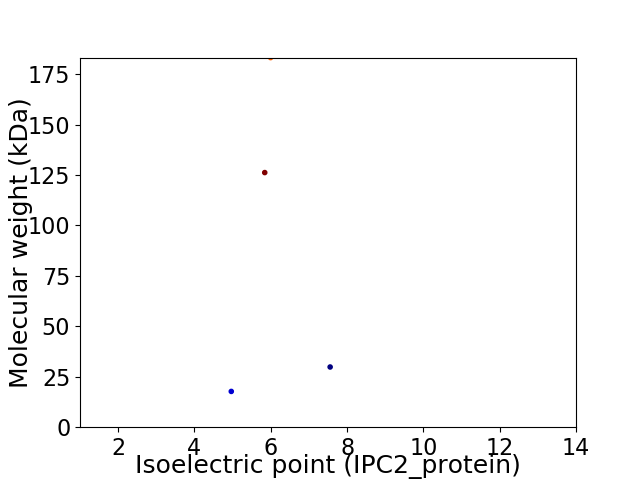
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|T1WEZ3|T1WEZ3_9VIRU Capsid protein OS=Tomato mottle mosaic virus OX=1391702 GN=CP PE=3 SV=1
MM1 pKa = 7.66SYY3 pKa = 10.99AITSPSQFVFLSSAWADD20 pKa = 3.29PVEE23 pKa = 5.21LINICTNSLGNQFQTQQARR42 pKa = 11.84TTVQQQFSEE51 pKa = 4.29VWKK54 pKa = 9.34PFPQSTVRR62 pKa = 11.84FPDD65 pKa = 3.24NVFKK69 pKa = 10.86VYY71 pKa = 10.34RR72 pKa = 11.84YY73 pKa = 9.93NAVIDD78 pKa = 4.13PLITALLGTFDD89 pKa = 3.24TRR91 pKa = 11.84NRR93 pKa = 11.84IIEE96 pKa = 4.17VEE98 pKa = 4.14NQQSPTTAEE107 pKa = 3.94TLDD110 pKa = 3.11ATRR113 pKa = 11.84RR114 pKa = 11.84VDD116 pKa = 3.5DD117 pKa = 3.56ATVAIRR123 pKa = 11.84SAVNNLVNEE132 pKa = 4.59LVRR135 pKa = 11.84GTGFYY140 pKa = 10.4NQSTFEE146 pKa = 4.24SMSGLAWTSAPASS159 pKa = 3.64
MM1 pKa = 7.66SYY3 pKa = 10.99AITSPSQFVFLSSAWADD20 pKa = 3.29PVEE23 pKa = 5.21LINICTNSLGNQFQTQQARR42 pKa = 11.84TTVQQQFSEE51 pKa = 4.29VWKK54 pKa = 9.34PFPQSTVRR62 pKa = 11.84FPDD65 pKa = 3.24NVFKK69 pKa = 10.86VYY71 pKa = 10.34RR72 pKa = 11.84YY73 pKa = 9.93NAVIDD78 pKa = 4.13PLITALLGTFDD89 pKa = 3.24TRR91 pKa = 11.84NRR93 pKa = 11.84IIEE96 pKa = 4.17VEE98 pKa = 4.14NQQSPTTAEE107 pKa = 3.94TLDD110 pKa = 3.11ATRR113 pKa = 11.84RR114 pKa = 11.84VDD116 pKa = 3.5DD117 pKa = 3.56ATVAIRR123 pKa = 11.84SAVNNLVNEE132 pKa = 4.59LVRR135 pKa = 11.84GTGFYY140 pKa = 10.4NQSTFEE146 pKa = 4.24SMSGLAWTSAPASS159 pKa = 3.64
Molecular weight: 17.74 kDa
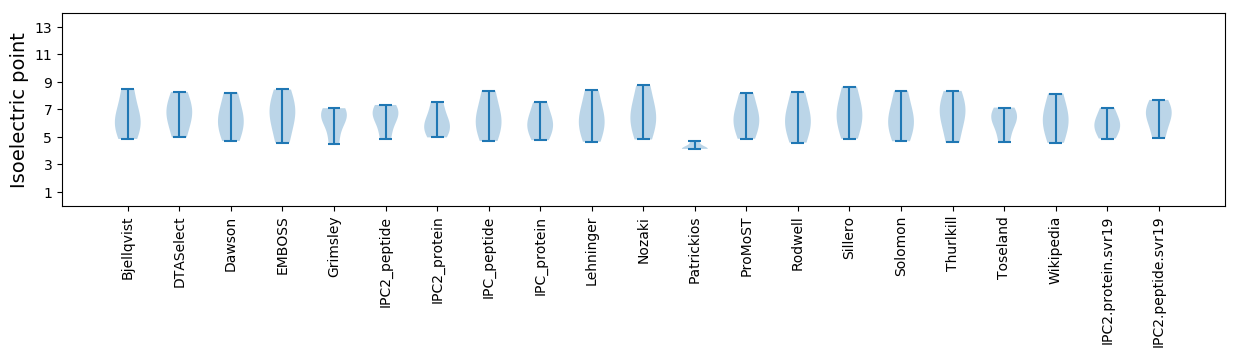
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|T1WEP4|T1WEP4_9VIRU Methyltransferase/RNA helicase OS=Tomato mottle mosaic virus OX=1391702 PE=4 SV=1
MM1 pKa = 7.68ALTVSGKK8 pKa = 9.44VRR10 pKa = 11.84ISEE13 pKa = 4.76FIDD16 pKa = 3.58LSKK19 pKa = 10.87SEE21 pKa = 4.5RR22 pKa = 11.84LLPSMFTHH30 pKa = 6.09VKK32 pKa = 9.38SVSVSKK38 pKa = 10.72VDD40 pKa = 3.01KK41 pKa = 11.32VMVNEE46 pKa = 4.12EE47 pKa = 4.31DD48 pKa = 3.7SLSEE52 pKa = 4.08VNLLKK57 pKa = 10.54GVKK60 pKa = 9.87LIDD63 pKa = 3.69GGYY66 pKa = 8.67VCLAGLVVSGEE77 pKa = 3.84WNLPDD82 pKa = 3.43NCRR85 pKa = 11.84GGVSICLVDD94 pKa = 4.74KK95 pKa = 10.99RR96 pKa = 11.84MQRR99 pKa = 11.84ADD101 pKa = 3.35EE102 pKa = 4.34ATLGSYY108 pKa = 7.82YY109 pKa = 10.45TGAAKK114 pKa = 10.18KK115 pKa = 9.96RR116 pKa = 11.84FQFKK120 pKa = 10.0IVPNYY125 pKa = 10.84AITTKK130 pKa = 10.61DD131 pKa = 2.95AEE133 pKa = 4.29RR134 pKa = 11.84NIWQVLVNIRR144 pKa = 11.84NVKK147 pKa = 8.31MAGGFCPLSLEE158 pKa = 4.22FVSVCIVYY166 pKa = 10.57KK167 pKa = 11.11NNIKK171 pKa = 10.61LGLRR175 pKa = 11.84EE176 pKa = 4.37KK177 pKa = 9.58ITRR180 pKa = 11.84VDD182 pKa = 3.31DD183 pKa = 4.44AGPIEE188 pKa = 4.28LTEE191 pKa = 4.08EE192 pKa = 4.25VVDD195 pKa = 4.03EE196 pKa = 4.6FMEE199 pKa = 4.38SVPMSVRR206 pKa = 11.84LAKK209 pKa = 10.42FRR211 pKa = 11.84TKK213 pKa = 10.38SSKK216 pKa = 10.35RR217 pKa = 11.84GPKK220 pKa = 9.83HH221 pKa = 6.05NSNNTNEE228 pKa = 4.23RR229 pKa = 11.84KK230 pKa = 9.91GRR232 pKa = 11.84SNFRR236 pKa = 11.84KK237 pKa = 10.11KK238 pKa = 10.07QDD240 pKa = 3.33QEE242 pKa = 4.68SYY244 pKa = 11.0GVSDD248 pKa = 3.99SLDD251 pKa = 3.41NLIEE255 pKa = 5.46DD256 pKa = 4.03DD257 pKa = 4.44TEE259 pKa = 4.3TSVAGSDD266 pKa = 3.75SYY268 pKa = 12.17
MM1 pKa = 7.68ALTVSGKK8 pKa = 9.44VRR10 pKa = 11.84ISEE13 pKa = 4.76FIDD16 pKa = 3.58LSKK19 pKa = 10.87SEE21 pKa = 4.5RR22 pKa = 11.84LLPSMFTHH30 pKa = 6.09VKK32 pKa = 9.38SVSVSKK38 pKa = 10.72VDD40 pKa = 3.01KK41 pKa = 11.32VMVNEE46 pKa = 4.12EE47 pKa = 4.31DD48 pKa = 3.7SLSEE52 pKa = 4.08VNLLKK57 pKa = 10.54GVKK60 pKa = 9.87LIDD63 pKa = 3.69GGYY66 pKa = 8.67VCLAGLVVSGEE77 pKa = 3.84WNLPDD82 pKa = 3.43NCRR85 pKa = 11.84GGVSICLVDD94 pKa = 4.74KK95 pKa = 10.99RR96 pKa = 11.84MQRR99 pKa = 11.84ADD101 pKa = 3.35EE102 pKa = 4.34ATLGSYY108 pKa = 7.82YY109 pKa = 10.45TGAAKK114 pKa = 10.18KK115 pKa = 9.96RR116 pKa = 11.84FQFKK120 pKa = 10.0IVPNYY125 pKa = 10.84AITTKK130 pKa = 10.61DD131 pKa = 2.95AEE133 pKa = 4.29RR134 pKa = 11.84NIWQVLVNIRR144 pKa = 11.84NVKK147 pKa = 8.31MAGGFCPLSLEE158 pKa = 4.22FVSVCIVYY166 pKa = 10.57KK167 pKa = 11.11NNIKK171 pKa = 10.61LGLRR175 pKa = 11.84EE176 pKa = 4.37KK177 pKa = 9.58ITRR180 pKa = 11.84VDD182 pKa = 3.31DD183 pKa = 4.44AGPIEE188 pKa = 4.28LTEE191 pKa = 4.08EE192 pKa = 4.25VVDD195 pKa = 4.03EE196 pKa = 4.6FMEE199 pKa = 4.38SVPMSVRR206 pKa = 11.84LAKK209 pKa = 10.42FRR211 pKa = 11.84TKK213 pKa = 10.38SSKK216 pKa = 10.35RR217 pKa = 11.84GPKK220 pKa = 9.83HH221 pKa = 6.05NSNNTNEE228 pKa = 4.23RR229 pKa = 11.84KK230 pKa = 9.91GRR232 pKa = 11.84SNFRR236 pKa = 11.84KK237 pKa = 10.11KK238 pKa = 10.07QDD240 pKa = 3.33QEE242 pKa = 4.68SYY244 pKa = 11.0GVSDD248 pKa = 3.99SLDD251 pKa = 3.41NLIEE255 pKa = 5.46DD256 pKa = 4.03DD257 pKa = 4.44TEE259 pKa = 4.3TSVAGSDD266 pKa = 3.75SYY268 pKa = 12.17
Molecular weight: 29.85 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3159 |
159 |
1616 |
789.8 |
89.27 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.648 ± 0.456 | 1.773 ± 0.179 |
6.046 ± 0.304 | 6.299 ± 0.342 |
4.843 ± 0.337 | 4.4 ± 0.438 |
2.089 ± 0.46 | 4.78 ± 0.064 |
6.394 ± 0.794 | 9.528 ± 0.618 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.722 ± 0.225 | 4.179 ± 0.607 |
3.292 ± 0.262 | 3.45 ± 0.602 |
5.192 ± 0.243 | 8.452 ± 0.419 |
6.141 ± 0.679 | 9.022 ± 0.457 |
0.855 ± 0.154 | 3.862 ± 0.359 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
