
Sphingobacteriales bacterium UPWRP_1
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Sphingobacteriia; Sphingobacteriales; unclassified Sphingobacteriales
Average proteome isoelectric point is 6.67
Get precalculated fractions of proteins
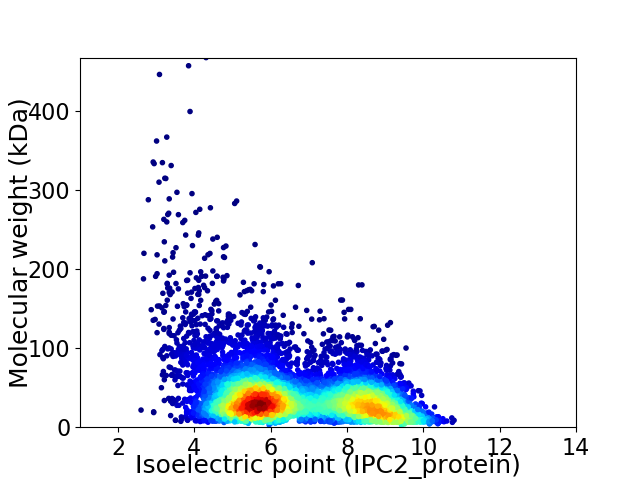
Virtual 2D-PAGE plot for 7781 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
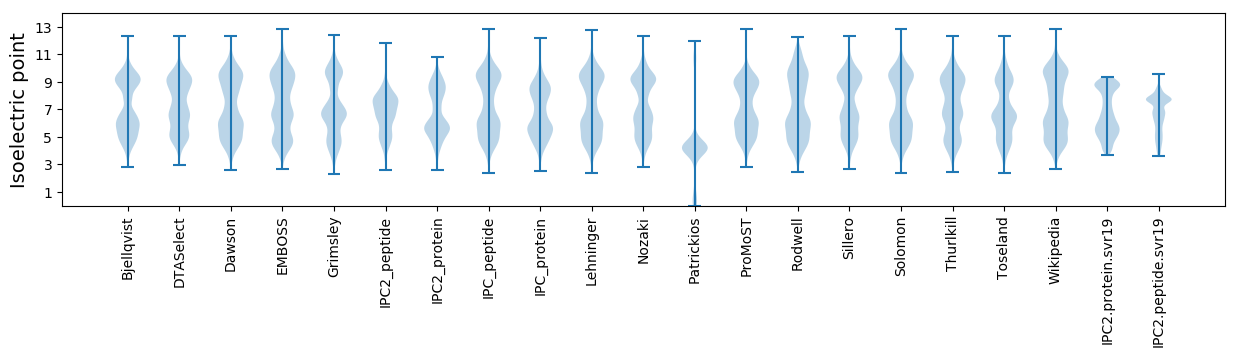
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2P7THF5|A0A2P7THF5_9SPHI Peptidase_M1 domain-containing protein OS=Sphingobacteriales bacterium UPWRP_1 OX=2116516 GN=C7N43_25535 PE=4 SV=1
MM1 pKa = 7.72KK2 pKa = 9.36KK3 pKa = 9.06TIFLSTFLLITALYY17 pKa = 9.82DD18 pKa = 4.16LKK20 pKa = 11.02AQNWTQIGVDD30 pKa = 3.1IDD32 pKa = 5.34GEE34 pKa = 4.6TEE36 pKa = 4.38DD37 pKa = 3.55NWSGYY42 pKa = 8.7SVSLSANGNIVAIGEE57 pKa = 4.33PLTDD61 pKa = 3.28EE62 pKa = 4.36TGIDD66 pKa = 3.65DD67 pKa = 4.12GQVRR71 pKa = 11.84VYY73 pKa = 10.92QNNDD77 pKa = 3.34GNWTQIGSDD86 pKa = 3.19IVGEE90 pKa = 4.04AAGDD94 pKa = 3.69RR95 pKa = 11.84FGSAVSLSAGGDD107 pKa = 3.19IVAVSAPRR115 pKa = 11.84NDD117 pKa = 4.02GNGTDD122 pKa = 3.49AGHH125 pKa = 5.62VRR127 pKa = 11.84VYY129 pKa = 11.16QNVSGNWTQIGQDD142 pKa = 3.09IDD144 pKa = 3.6GQAADD149 pKa = 4.88DD150 pKa = 3.98RR151 pKa = 11.84SGDD154 pKa = 3.87AVSLSANGSILAIGSVRR171 pKa = 11.84NEE173 pKa = 3.2AWAGDD178 pKa = 3.3VRR180 pKa = 11.84VYY182 pKa = 11.24QNVSGNWTQIGSDD195 pKa = 2.82IVGEE199 pKa = 4.18NPSDD203 pKa = 3.46QSGYY207 pKa = 10.46SVSLNATGNILAIGAFANSDD227 pKa = 3.52NGNLAGGQVRR237 pKa = 11.84VYY239 pKa = 11.1QNVSGNWTQVGQDD252 pKa = 2.91INGYY256 pKa = 7.59FQEE259 pKa = 4.19NLLGYY264 pKa = 9.72SVSLNATGNILAIGAPGVNAAGFAQVFQNISGTWTQIGEE303 pKa = 4.45DD304 pKa = 3.38IYY306 pKa = 11.65GEE308 pKa = 4.03NDD310 pKa = 3.06FDD312 pKa = 4.56EE313 pKa = 5.23SGCSVSLNANGNIVAIGSRR332 pKa = 11.84GVEE335 pKa = 4.12GIGNIDD341 pKa = 3.31GSVRR345 pKa = 11.84VYY347 pKa = 11.26EE348 pKa = 4.21NVSGSWLQTGNTIAGEE364 pKa = 4.08PLNQFPGIAVSLNAGGNILAIGAPYY389 pKa = 10.8NNGNGEE395 pKa = 4.16EE396 pKa = 4.1AGHH399 pKa = 5.3VRR401 pKa = 11.84VYY403 pKa = 7.84QQCDD407 pKa = 3.04INTPPVPTIATLPDD421 pKa = 3.2VTAEE425 pKa = 4.16CSVTTLTPPTATDD438 pKa = 3.45GCGNTVFGTPSVTLPLTSQGTTTVIWIYY466 pKa = 11.2NSGNASSVQTQNVVIDD482 pKa = 4.44DD483 pKa = 3.93VTNPTITCVGNQTVDD498 pKa = 2.91ADD500 pKa = 3.51QSHH503 pKa = 7.38FYY505 pKa = 9.08TVNGTEE511 pKa = 4.39FDD513 pKa = 3.75PTLTSDD519 pKa = 3.14NCGIASVINLYY530 pKa = 8.87TVAFSLAGAQIPEE543 pKa = 4.34GNTTISWTITDD554 pKa = 3.58NAGNNQTCSFVVTVNTYY571 pKa = 10.71VGIEE575 pKa = 3.96TLQQKK580 pKa = 10.28GISIYY585 pKa = 10.2PNPANDD591 pKa = 3.45ILHH594 pKa = 7.2IDD596 pKa = 4.36FAQNNIQKK604 pKa = 10.44LAIKK608 pKa = 9.43DD609 pKa = 3.52IKK611 pKa = 10.68GSSIFEE617 pKa = 4.06KK618 pKa = 10.38TNPNQNEE625 pKa = 4.19TLDD628 pKa = 4.05LSDD631 pKa = 4.72FASGMYY637 pKa = 10.05IMSIQTDD644 pKa = 3.84KK645 pKa = 11.26EE646 pKa = 3.81ILITKK651 pKa = 9.63IVKK654 pKa = 9.03QQ655 pKa = 3.63
MM1 pKa = 7.72KK2 pKa = 9.36KK3 pKa = 9.06TIFLSTFLLITALYY17 pKa = 9.82DD18 pKa = 4.16LKK20 pKa = 11.02AQNWTQIGVDD30 pKa = 3.1IDD32 pKa = 5.34GEE34 pKa = 4.6TEE36 pKa = 4.38DD37 pKa = 3.55NWSGYY42 pKa = 8.7SVSLSANGNIVAIGEE57 pKa = 4.33PLTDD61 pKa = 3.28EE62 pKa = 4.36TGIDD66 pKa = 3.65DD67 pKa = 4.12GQVRR71 pKa = 11.84VYY73 pKa = 10.92QNNDD77 pKa = 3.34GNWTQIGSDD86 pKa = 3.19IVGEE90 pKa = 4.04AAGDD94 pKa = 3.69RR95 pKa = 11.84FGSAVSLSAGGDD107 pKa = 3.19IVAVSAPRR115 pKa = 11.84NDD117 pKa = 4.02GNGTDD122 pKa = 3.49AGHH125 pKa = 5.62VRR127 pKa = 11.84VYY129 pKa = 11.16QNVSGNWTQIGQDD142 pKa = 3.09IDD144 pKa = 3.6GQAADD149 pKa = 4.88DD150 pKa = 3.98RR151 pKa = 11.84SGDD154 pKa = 3.87AVSLSANGSILAIGSVRR171 pKa = 11.84NEE173 pKa = 3.2AWAGDD178 pKa = 3.3VRR180 pKa = 11.84VYY182 pKa = 11.24QNVSGNWTQIGSDD195 pKa = 2.82IVGEE199 pKa = 4.18NPSDD203 pKa = 3.46QSGYY207 pKa = 10.46SVSLNATGNILAIGAFANSDD227 pKa = 3.52NGNLAGGQVRR237 pKa = 11.84VYY239 pKa = 11.1QNVSGNWTQVGQDD252 pKa = 2.91INGYY256 pKa = 7.59FQEE259 pKa = 4.19NLLGYY264 pKa = 9.72SVSLNATGNILAIGAPGVNAAGFAQVFQNISGTWTQIGEE303 pKa = 4.45DD304 pKa = 3.38IYY306 pKa = 11.65GEE308 pKa = 4.03NDD310 pKa = 3.06FDD312 pKa = 4.56EE313 pKa = 5.23SGCSVSLNANGNIVAIGSRR332 pKa = 11.84GVEE335 pKa = 4.12GIGNIDD341 pKa = 3.31GSVRR345 pKa = 11.84VYY347 pKa = 11.26EE348 pKa = 4.21NVSGSWLQTGNTIAGEE364 pKa = 4.08PLNQFPGIAVSLNAGGNILAIGAPYY389 pKa = 10.8NNGNGEE395 pKa = 4.16EE396 pKa = 4.1AGHH399 pKa = 5.3VRR401 pKa = 11.84VYY403 pKa = 7.84QQCDD407 pKa = 3.04INTPPVPTIATLPDD421 pKa = 3.2VTAEE425 pKa = 4.16CSVTTLTPPTATDD438 pKa = 3.45GCGNTVFGTPSVTLPLTSQGTTTVIWIYY466 pKa = 11.2NSGNASSVQTQNVVIDD482 pKa = 4.44DD483 pKa = 3.93VTNPTITCVGNQTVDD498 pKa = 2.91ADD500 pKa = 3.51QSHH503 pKa = 7.38FYY505 pKa = 9.08TVNGTEE511 pKa = 4.39FDD513 pKa = 3.75PTLTSDD519 pKa = 3.14NCGIASVINLYY530 pKa = 8.87TVAFSLAGAQIPEE543 pKa = 4.34GNTTISWTITDD554 pKa = 3.58NAGNNQTCSFVVTVNTYY571 pKa = 10.71VGIEE575 pKa = 3.96TLQQKK580 pKa = 10.28GISIYY585 pKa = 10.2PNPANDD591 pKa = 3.45ILHH594 pKa = 7.2IDD596 pKa = 4.36FAQNNIQKK604 pKa = 10.44LAIKK608 pKa = 9.43DD609 pKa = 3.52IKK611 pKa = 10.68GSSIFEE617 pKa = 4.06KK618 pKa = 10.38TNPNQNEE625 pKa = 4.19TLDD628 pKa = 4.05LSDD631 pKa = 4.72FASGMYY637 pKa = 10.05IMSIQTDD644 pKa = 3.84KK645 pKa = 11.26EE646 pKa = 3.81ILITKK651 pKa = 9.63IVKK654 pKa = 9.03QQ655 pKa = 3.63
Molecular weight: 68.69 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2P7TI51|A0A2P7TI51_9SPHI PKD domain-containing protein (Fragment) OS=Sphingobacteriales bacterium UPWRP_1 OX=2116516 GN=C7N43_24245 PE=4 SV=1
MM1 pKa = 7.4AHH3 pKa = 7.28PKK5 pKa = 9.68RR6 pKa = 11.84RR7 pKa = 11.84HH8 pKa = 5.03SKK10 pKa = 6.87TRR12 pKa = 11.84RR13 pKa = 11.84DD14 pKa = 3.28KK15 pKa = 11.26RR16 pKa = 11.84RR17 pKa = 11.84THH19 pKa = 5.36YY20 pKa = 8.65TASVPNVVNCSNCGAPVLSHH40 pKa = 6.49RR41 pKa = 11.84VCGEE45 pKa = 3.63CGFYY49 pKa = 10.38RR50 pKa = 11.84GQLAIAKK57 pKa = 6.44TTAA60 pKa = 2.97
MM1 pKa = 7.4AHH3 pKa = 7.28PKK5 pKa = 9.68RR6 pKa = 11.84RR7 pKa = 11.84HH8 pKa = 5.03SKK10 pKa = 6.87TRR12 pKa = 11.84RR13 pKa = 11.84DD14 pKa = 3.28KK15 pKa = 11.26RR16 pKa = 11.84RR17 pKa = 11.84THH19 pKa = 5.36YY20 pKa = 8.65TASVPNVVNCSNCGAPVLSHH40 pKa = 6.49RR41 pKa = 11.84VCGEE45 pKa = 3.63CGFYY49 pKa = 10.38RR50 pKa = 11.84GQLAIAKK57 pKa = 6.44TTAA60 pKa = 2.97
Molecular weight: 6.7 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2977425 |
25 |
4588 |
382.7 |
42.42 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.435 ± 0.026 | 1.317 ± 0.018 |
4.668 ± 0.02 | 4.973 ± 0.036 |
4.739 ± 0.022 | 7.102 ± 0.035 |
1.847 ± 0.016 | 6.219 ± 0.023 |
5.003 ± 0.045 | 9.625 ± 0.042 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.061 ± 0.015 | 6.168 ± 0.031 |
4.777 ± 0.023 | 4.565 ± 0.023 |
3.599 ± 0.026 | 5.764 ± 0.032 |
7.209 ± 0.061 | 6.495 ± 0.023 |
1.239 ± 0.01 | 4.196 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
