
Rhizobium gallicum bv. gallicum R602
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Rhizobiaceae; Rhizobium/Agrobacterium group; Rhizobium; Rhizobium gallicum; Rhizobium gallicum bv. gallicum
Average proteome isoelectric point is 6.6
Get precalculated fractions of proteins
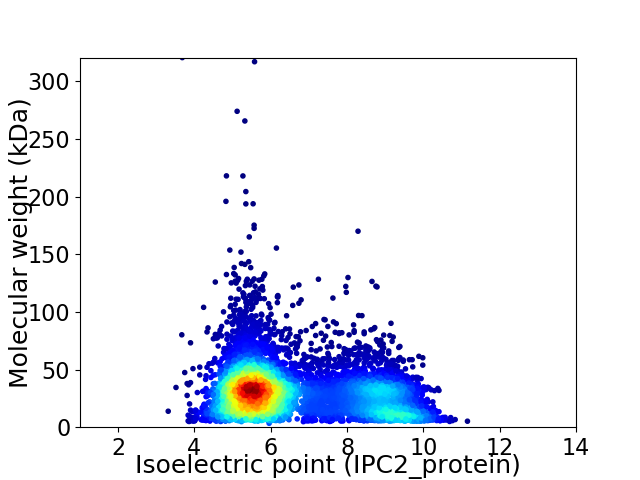
Virtual 2D-PAGE plot for 7034 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
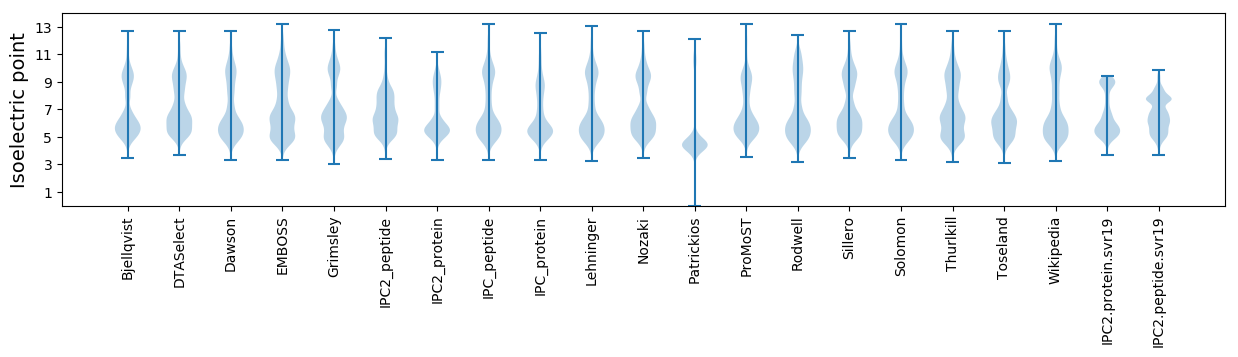
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0B4X786|A0A0B4X786_9RHIZ CbtA superfamily transporter protein OS=Rhizobium gallicum bv. gallicum R602 OX=1418105 GN=RGR602_CH03280 PE=4 SV=1
MM1 pKa = 7.38ATLTVNADD9 pKa = 3.18YY10 pKa = 11.16GLDD13 pKa = 3.28MRR15 pKa = 11.84GIDD18 pKa = 5.13FGWLPYY24 pKa = 10.39ADD26 pKa = 4.12SYY28 pKa = 11.56TYY30 pKa = 11.18GSTIFAAHH38 pKa = 6.63FSDD41 pKa = 3.68GTVEE45 pKa = 4.02EE46 pKa = 4.3FRR48 pKa = 11.84GRR50 pKa = 11.84GFSYY54 pKa = 10.87DD55 pKa = 3.28SDD57 pKa = 3.98GVPTGGTVRR66 pKa = 11.84SYY68 pKa = 12.04ALFDD72 pKa = 3.38NGNRR76 pKa = 11.84VSYY79 pKa = 11.23VDD81 pKa = 3.69GVSIAVTSIVDD92 pKa = 3.29AAEE95 pKa = 4.41TYY97 pKa = 7.57TTSDD101 pKa = 4.12DD102 pKa = 3.28IAVIGRR108 pKa = 11.84ALAGNDD114 pKa = 3.24VLRR117 pKa = 11.84GGGIFDD123 pKa = 4.06YY124 pKa = 11.24LRR126 pKa = 11.84GFAGNDD132 pKa = 3.3TLYY135 pKa = 11.42GNAGNDD141 pKa = 3.62TLVGDD146 pKa = 5.22DD147 pKa = 5.25GNDD150 pKa = 3.91TINGGAGKK158 pKa = 10.12DD159 pKa = 3.59RR160 pKa = 11.84IDD162 pKa = 3.92GGVGSDD168 pKa = 3.1TASYY172 pKa = 10.68AGASSGVVASLGNPGGNTQNAYY194 pKa = 9.97GDD196 pKa = 3.64IYY198 pKa = 10.87ISIEE202 pKa = 3.99SLAGSSHH209 pKa = 7.4ADD211 pKa = 3.14RR212 pKa = 11.84LYY214 pKa = 11.47GNAATNYY221 pKa = 10.14LSGDD225 pKa = 3.3AGNDD229 pKa = 3.37YY230 pKa = 11.15LSASGGNDD238 pKa = 2.88WLHH241 pKa = 6.5GGSGADD247 pKa = 3.3QLVGGSGADD256 pKa = 3.21RR257 pKa = 11.84FIFKK261 pKa = 10.37AWGDD265 pKa = 3.64HH266 pKa = 6.29SGDD269 pKa = 4.06SILDD273 pKa = 3.56FTATQSDD280 pKa = 4.49RR281 pKa = 11.84IDD283 pKa = 3.45MSKK286 pKa = 10.07IDD288 pKa = 4.41ANQSIAGDD296 pKa = 3.52QAFSFIGTAAFSGKK310 pKa = 10.12AGEE313 pKa = 4.09LRR315 pKa = 11.84FVKK318 pKa = 9.53TASDD322 pKa = 3.56TYY324 pKa = 10.87IYY326 pKa = 11.1GDD328 pKa = 3.63VNGDD332 pKa = 2.99ASTDD336 pKa = 3.72YY337 pKa = 10.73TIHH340 pKa = 7.42LDD342 pKa = 3.93DD343 pKa = 6.12AVTIWNSYY351 pKa = 10.17FYY353 pKa = 11.29LL354 pKa = 5.37
MM1 pKa = 7.38ATLTVNADD9 pKa = 3.18YY10 pKa = 11.16GLDD13 pKa = 3.28MRR15 pKa = 11.84GIDD18 pKa = 5.13FGWLPYY24 pKa = 10.39ADD26 pKa = 4.12SYY28 pKa = 11.56TYY30 pKa = 11.18GSTIFAAHH38 pKa = 6.63FSDD41 pKa = 3.68GTVEE45 pKa = 4.02EE46 pKa = 4.3FRR48 pKa = 11.84GRR50 pKa = 11.84GFSYY54 pKa = 10.87DD55 pKa = 3.28SDD57 pKa = 3.98GVPTGGTVRR66 pKa = 11.84SYY68 pKa = 12.04ALFDD72 pKa = 3.38NGNRR76 pKa = 11.84VSYY79 pKa = 11.23VDD81 pKa = 3.69GVSIAVTSIVDD92 pKa = 3.29AAEE95 pKa = 4.41TYY97 pKa = 7.57TTSDD101 pKa = 4.12DD102 pKa = 3.28IAVIGRR108 pKa = 11.84ALAGNDD114 pKa = 3.24VLRR117 pKa = 11.84GGGIFDD123 pKa = 4.06YY124 pKa = 11.24LRR126 pKa = 11.84GFAGNDD132 pKa = 3.3TLYY135 pKa = 11.42GNAGNDD141 pKa = 3.62TLVGDD146 pKa = 5.22DD147 pKa = 5.25GNDD150 pKa = 3.91TINGGAGKK158 pKa = 10.12DD159 pKa = 3.59RR160 pKa = 11.84IDD162 pKa = 3.92GGVGSDD168 pKa = 3.1TASYY172 pKa = 10.68AGASSGVVASLGNPGGNTQNAYY194 pKa = 9.97GDD196 pKa = 3.64IYY198 pKa = 10.87ISIEE202 pKa = 3.99SLAGSSHH209 pKa = 7.4ADD211 pKa = 3.14RR212 pKa = 11.84LYY214 pKa = 11.47GNAATNYY221 pKa = 10.14LSGDD225 pKa = 3.3AGNDD229 pKa = 3.37YY230 pKa = 11.15LSASGGNDD238 pKa = 2.88WLHH241 pKa = 6.5GGSGADD247 pKa = 3.3QLVGGSGADD256 pKa = 3.21RR257 pKa = 11.84FIFKK261 pKa = 10.37AWGDD265 pKa = 3.64HH266 pKa = 6.29SGDD269 pKa = 4.06SILDD273 pKa = 3.56FTATQSDD280 pKa = 4.49RR281 pKa = 11.84IDD283 pKa = 3.45MSKK286 pKa = 10.07IDD288 pKa = 4.41ANQSIAGDD296 pKa = 3.52QAFSFIGTAAFSGKK310 pKa = 10.12AGEE313 pKa = 4.09LRR315 pKa = 11.84FVKK318 pKa = 9.53TASDD322 pKa = 3.56TYY324 pKa = 10.87IYY326 pKa = 11.1GDD328 pKa = 3.63VNGDD332 pKa = 2.99ASTDD336 pKa = 3.72YY337 pKa = 10.73TIHH340 pKa = 7.42LDD342 pKa = 3.93DD343 pKa = 6.12AVTIWNSYY351 pKa = 10.17FYY353 pKa = 11.29LL354 pKa = 5.37
Molecular weight: 36.8 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0B4WWL8|A0A0B4WWL8_9RHIZ Phosphoenolpyruvate carboxykinase (ATP) OS=Rhizobium gallicum bv. gallicum R602 OX=1418105 GN=pckA PE=3 SV=1
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.73LVRR12 pKa = 11.84KK13 pKa = 9.15RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.1GGRR28 pKa = 11.84KK29 pKa = 9.65VIVARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.03RR41 pKa = 11.84LSAA44 pKa = 4.03
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.73LVRR12 pKa = 11.84KK13 pKa = 9.15RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.1GGRR28 pKa = 11.84KK29 pKa = 9.65VIVARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.03RR41 pKa = 11.84LSAA44 pKa = 4.03
Molecular weight: 5.19 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2066382 |
29 |
3168 |
293.8 |
32.03 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.746 ± 0.037 | 0.881 ± 0.01 |
5.575 ± 0.022 | 5.789 ± 0.023 |
3.97 ± 0.02 | 8.191 ± 0.025 |
2.053 ± 0.015 | 5.779 ± 0.02 |
3.793 ± 0.026 | 9.921 ± 0.031 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.57 ± 0.014 | 2.891 ± 0.015 |
4.886 ± 0.023 | 3.121 ± 0.019 |
6.752 ± 0.028 | 5.922 ± 0.02 |
5.233 ± 0.02 | 7.306 ± 0.025 |
1.283 ± 0.012 | 2.336 ± 0.013 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
