
Peptidiphaga gingivicola
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Actinomycetales; Actinomycetaceae; Peptidiphaga
Average proteome isoelectric point is 6.34
Get precalculated fractions of proteins
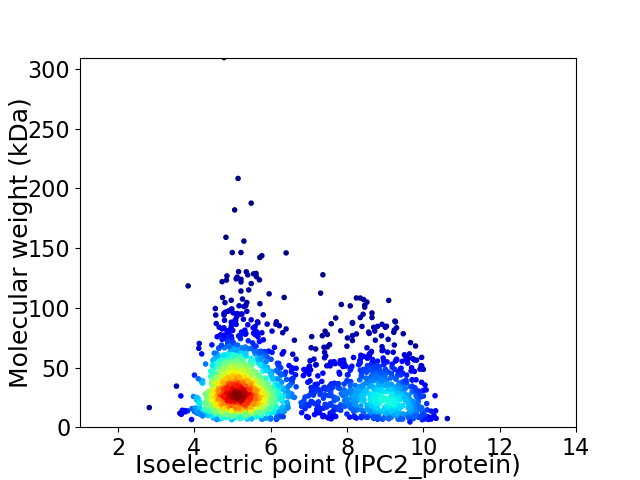
Virtual 2D-PAGE plot for 1994 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
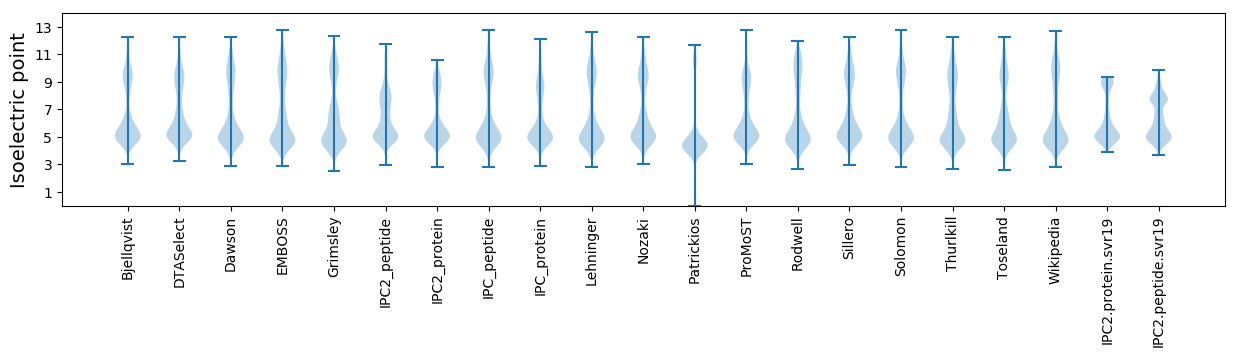
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A179B561|A0A179B561_9ACTO BAAT_C domain-containing protein OS=Peptidiphaga gingivicola OX=2741497 GN=A4H34_06905 PE=4 SV=1
MM1 pKa = 6.93YY2 pKa = 10.46ASIAASFRR10 pKa = 11.84AAIFSSILTKK20 pKa = 10.74SSGVDD25 pKa = 3.3VEE27 pKa = 4.65FTMALNTPCGVVSAYY42 pKa = 11.04SFILLYY48 pKa = 10.44IFNQVKK54 pKa = 10.35DD55 pKa = 3.87VGLGQPPNAEE65 pKa = 4.14MLDD68 pKa = 3.77MPLLLIGIVMEE79 pKa = 4.28STTFSNQDD87 pKa = 2.78SS88 pKa = 3.72
MM1 pKa = 6.93YY2 pKa = 10.46ASIAASFRR10 pKa = 11.84AAIFSSILTKK20 pKa = 10.74SSGVDD25 pKa = 3.3VEE27 pKa = 4.65FTMALNTPCGVVSAYY42 pKa = 11.04SFILLYY48 pKa = 10.44IFNQVKK54 pKa = 10.35DD55 pKa = 3.87VGLGQPPNAEE65 pKa = 4.14MLDD68 pKa = 3.77MPLLLIGIVMEE79 pKa = 4.28STTFSNQDD87 pKa = 2.78SS88 pKa = 3.72
Molecular weight: 9.46 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A179B5X1|A0A179B5X1_9ACTO Ribonuclease OS=Peptidiphaga gingivicola OX=2741497 GN=A4H34_06610 PE=4 SV=1
MM1 pKa = 7.27TALDD5 pKa = 4.01NLVMTFALPDD15 pKa = 3.37IKK17 pKa = 10.68RR18 pKa = 11.84EE19 pKa = 3.94LGATVTQLQWFVNAYY34 pKa = 7.34TLVVASALLPMAALGDD50 pKa = 3.59RR51 pKa = 11.84FGRR54 pKa = 11.84RR55 pKa = 11.84RR56 pKa = 11.84IFSYY60 pKa = 10.68GIAVFAAASAACALAPTPEE79 pKa = 3.93ALIVARR85 pKa = 11.84AIQGAGGAAIVTLSLALLVDD105 pKa = 3.92AVPRR109 pKa = 11.84RR110 pKa = 11.84LRR112 pKa = 11.84EE113 pKa = 3.66LAIGVWGGVNGLGIAMGPLLGGAVVSGLHH142 pKa = 5.23WSVVFWINVPIGALSLPLVPRR163 pKa = 11.84FLPRR167 pKa = 11.84AAAGSKK173 pKa = 9.19GADD176 pKa = 3.23WIGTILGIGFVFPLTWAVVEE196 pKa = 4.67GPSRR200 pKa = 11.84GWTDD204 pKa = 2.94GLTIGGFAVAGTCLVLFLLWEE225 pKa = 4.73RR226 pKa = 11.84SAKK229 pKa = 10.63APIIPLSLFANRR241 pKa = 11.84RR242 pKa = 11.84FSLVNAATVLFAAGVFGAIFFLSQFLQITLGYY274 pKa = 9.58SAFEE278 pKa = 4.29AGLRR282 pKa = 11.84AGPWTLLPLFVSPASGGLVKK302 pKa = 10.67RR303 pKa = 11.84LGVRR307 pKa = 11.84RR308 pKa = 11.84VLVSGMFLQTVALAWFAVKK327 pKa = 10.47VGADD331 pKa = 3.58VPYY334 pKa = 9.9GDD336 pKa = 5.7CIAPMAVAGLGMGLSFAPLATGALQGVSADD366 pKa = 3.69RR367 pKa = 11.84RR368 pKa = 11.84AVASGVNSTLRR379 pKa = 11.84HH380 pKa = 5.65LGVAIGIAACTAIFTAHH397 pKa = 6.32GKK399 pKa = 9.26YY400 pKa = 10.57LPGQPFVDD408 pKa = 5.43GIKK411 pKa = 10.34PSLWLCAALLATATVCAEE429 pKa = 4.01RR430 pKa = 11.84SDD432 pKa = 3.6RR433 pKa = 11.84AARR436 pKa = 11.84RR437 pKa = 3.49
MM1 pKa = 7.27TALDD5 pKa = 4.01NLVMTFALPDD15 pKa = 3.37IKK17 pKa = 10.68RR18 pKa = 11.84EE19 pKa = 3.94LGATVTQLQWFVNAYY34 pKa = 7.34TLVVASALLPMAALGDD50 pKa = 3.59RR51 pKa = 11.84FGRR54 pKa = 11.84RR55 pKa = 11.84RR56 pKa = 11.84IFSYY60 pKa = 10.68GIAVFAAASAACALAPTPEE79 pKa = 3.93ALIVARR85 pKa = 11.84AIQGAGGAAIVTLSLALLVDD105 pKa = 3.92AVPRR109 pKa = 11.84RR110 pKa = 11.84LRR112 pKa = 11.84EE113 pKa = 3.66LAIGVWGGVNGLGIAMGPLLGGAVVSGLHH142 pKa = 5.23WSVVFWINVPIGALSLPLVPRR163 pKa = 11.84FLPRR167 pKa = 11.84AAAGSKK173 pKa = 9.19GADD176 pKa = 3.23WIGTILGIGFVFPLTWAVVEE196 pKa = 4.67GPSRR200 pKa = 11.84GWTDD204 pKa = 2.94GLTIGGFAVAGTCLVLFLLWEE225 pKa = 4.73RR226 pKa = 11.84SAKK229 pKa = 10.63APIIPLSLFANRR241 pKa = 11.84RR242 pKa = 11.84FSLVNAATVLFAAGVFGAIFFLSQFLQITLGYY274 pKa = 9.58SAFEE278 pKa = 4.29AGLRR282 pKa = 11.84AGPWTLLPLFVSPASGGLVKK302 pKa = 10.67RR303 pKa = 11.84LGVRR307 pKa = 11.84RR308 pKa = 11.84VLVSGMFLQTVALAWFAVKK327 pKa = 10.47VGADD331 pKa = 3.58VPYY334 pKa = 9.9GDD336 pKa = 5.7CIAPMAVAGLGMGLSFAPLATGALQGVSADD366 pKa = 3.69RR367 pKa = 11.84RR368 pKa = 11.84AVASGVNSTLRR379 pKa = 11.84HH380 pKa = 5.65LGVAIGIAACTAIFTAHH397 pKa = 6.32GKK399 pKa = 9.26YY400 pKa = 10.57LPGQPFVDD408 pKa = 5.43GIKK411 pKa = 10.34PSLWLCAALLATATVCAEE429 pKa = 4.01RR430 pKa = 11.84SDD432 pKa = 3.6RR433 pKa = 11.84AARR436 pKa = 11.84RR437 pKa = 3.49
Molecular weight: 45.34 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
685011 |
37 |
2948 |
343.5 |
36.84 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.891 ± 0.085 | 0.779 ± 0.016 |
5.93 ± 0.046 | 6.354 ± 0.06 |
3.157 ± 0.036 | 9.249 ± 0.053 |
1.746 ± 0.021 | 4.243 ± 0.043 |
3.593 ± 0.046 | 9.007 ± 0.057 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.038 ± 0.025 | 2.449 ± 0.031 |
5.132 ± 0.046 | 2.709 ± 0.029 |
7.104 ± 0.064 | 6.646 ± 0.053 |
5.202 ± 0.041 | 8.297 ± 0.052 |
1.333 ± 0.02 | 2.139 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
