
Elusimicrobium sp. An273
Taxonomy: cellular organisms; Bacteria; Elusimicrobia; Elusimicrobia; Elusimicrobiales; Elusimicrobiaceae; Elusimicrobium; unclassified Elusimicrobium
Average proteome isoelectric point is 7.15
Get precalculated fractions of proteins
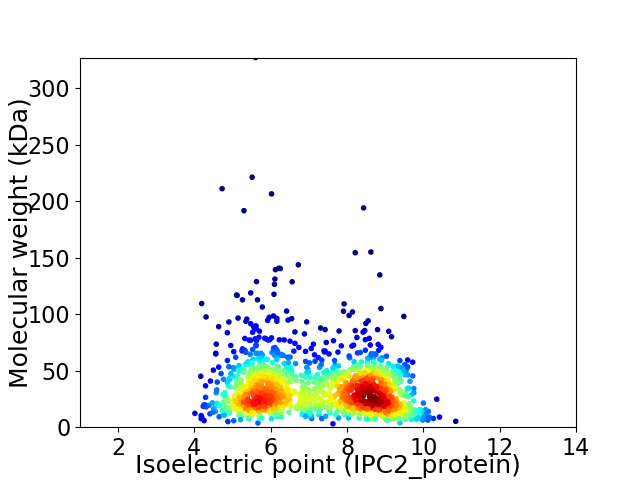
Virtual 2D-PAGE plot for 1395 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
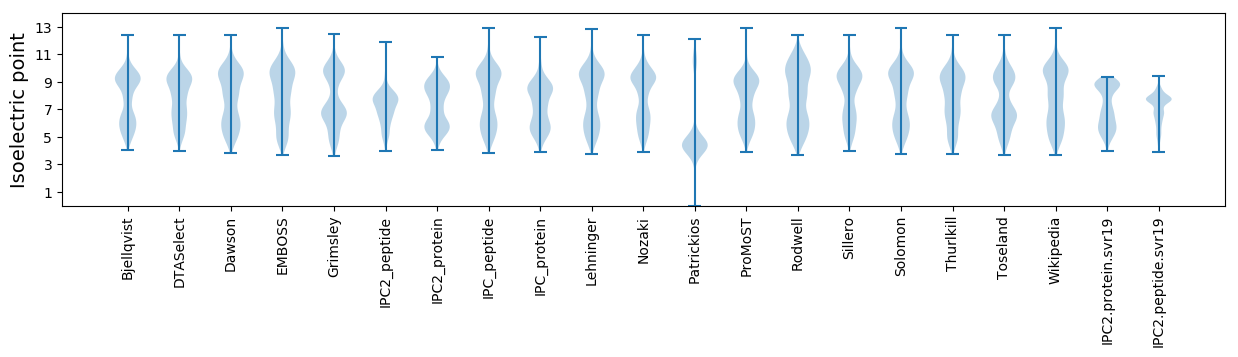
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1Y4DFR0|A0A1Y4DFR0_9BACT Uncharacterized protein OS=Elusimicrobium sp. An273 OX=1965617 GN=B5F75_04820 PE=4 SV=1
MM1 pKa = 7.18VKK3 pKa = 9.97YY4 pKa = 10.13VCEE7 pKa = 4.3VCGYY11 pKa = 9.03TYY13 pKa = 10.98DD14 pKa = 4.11PAVGDD19 pKa = 4.17PDD21 pKa = 3.73NGVPAGTKK29 pKa = 9.33WEE31 pKa = 4.16DD32 pKa = 3.39VKK34 pKa = 11.27EE35 pKa = 4.13DD36 pKa = 3.72WVCPVCGVGKK46 pKa = 10.01DD47 pKa = 3.4QFKK50 pKa = 10.96PEE52 pKa = 3.84NN53 pKa = 3.58
MM1 pKa = 7.18VKK3 pKa = 9.97YY4 pKa = 10.13VCEE7 pKa = 4.3VCGYY11 pKa = 9.03TYY13 pKa = 10.98DD14 pKa = 4.11PAVGDD19 pKa = 4.17PDD21 pKa = 3.73NGVPAGTKK29 pKa = 9.33WEE31 pKa = 4.16DD32 pKa = 3.39VKK34 pKa = 11.27EE35 pKa = 4.13DD36 pKa = 3.72WVCPVCGVGKK46 pKa = 10.01DD47 pKa = 3.4QFKK50 pKa = 10.96PEE52 pKa = 3.84NN53 pKa = 3.58
Molecular weight: 5.84 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1Y4DES5|A0A1Y4DES5_9BACT Uncharacterized protein OS=Elusimicrobium sp. An273 OX=1965617 GN=B5F75_02545 PE=4 SV=1
MM1 pKa = 7.59KK2 pKa = 9.08KK3 pKa = 8.18TLKK6 pKa = 9.71TLWRR10 pKa = 11.84LLLHH14 pKa = 6.44FFFPRR19 pKa = 11.84TCFACGCDD27 pKa = 3.73LPWRR31 pKa = 11.84RR32 pKa = 11.84EE33 pKa = 3.94EE34 pKa = 4.21PLCAACAQGLKK45 pKa = 9.58TPGPHH50 pKa = 5.34ICRR53 pKa = 11.84RR54 pKa = 11.84CGAVLTEE61 pKa = 5.09GGAHH65 pKa = 7.15CYY67 pKa = 10.03ACRR70 pKa = 11.84GSKK73 pKa = 9.3EE74 pKa = 3.57RR75 pKa = 11.84AYY77 pKa = 9.31KK78 pKa = 10.5CRR80 pKa = 11.84VIRR83 pKa = 11.84SAFVFNTSSRR93 pKa = 11.84ALVHH97 pKa = 5.74ALKK100 pKa = 10.9YY101 pKa = 10.77AGADD105 pKa = 3.24YY106 pKa = 10.46LARR109 pKa = 11.84YY110 pKa = 8.1MGGLMAKK117 pKa = 10.0RR118 pKa = 11.84FARR121 pKa = 11.84YY122 pKa = 8.87PEE124 pKa = 3.93LAAAEE129 pKa = 4.29VVMPVPLFPRR139 pKa = 11.84RR140 pKa = 11.84GRR142 pKa = 11.84KK143 pKa = 8.94RR144 pKa = 11.84GYY146 pKa = 9.28NQSEE150 pKa = 3.76LLARR154 pKa = 11.84FFAPQVGLPVQTKK167 pKa = 9.61ALVRR171 pKa = 11.84VRR173 pKa = 11.84NTVSQTKK180 pKa = 10.13LGRR183 pKa = 11.84AGRR186 pKa = 11.84LQNMAGAFACRR197 pKa = 11.84RR198 pKa = 11.84PEE200 pKa = 3.83WVKK203 pKa = 11.0GKK205 pKa = 8.03TVLLIDD211 pKa = 4.8DD212 pKa = 4.2VATTGATLEE221 pKa = 4.37GCAAALKK228 pKa = 10.27AAGARR233 pKa = 11.84RR234 pKa = 11.84VLAYY238 pKa = 9.2TFARR242 pKa = 11.84EE243 pKa = 3.86
MM1 pKa = 7.59KK2 pKa = 9.08KK3 pKa = 8.18TLKK6 pKa = 9.71TLWRR10 pKa = 11.84LLLHH14 pKa = 6.44FFFPRR19 pKa = 11.84TCFACGCDD27 pKa = 3.73LPWRR31 pKa = 11.84RR32 pKa = 11.84EE33 pKa = 3.94EE34 pKa = 4.21PLCAACAQGLKK45 pKa = 9.58TPGPHH50 pKa = 5.34ICRR53 pKa = 11.84RR54 pKa = 11.84CGAVLTEE61 pKa = 5.09GGAHH65 pKa = 7.15CYY67 pKa = 10.03ACRR70 pKa = 11.84GSKK73 pKa = 9.3EE74 pKa = 3.57RR75 pKa = 11.84AYY77 pKa = 9.31KK78 pKa = 10.5CRR80 pKa = 11.84VIRR83 pKa = 11.84SAFVFNTSSRR93 pKa = 11.84ALVHH97 pKa = 5.74ALKK100 pKa = 10.9YY101 pKa = 10.77AGADD105 pKa = 3.24YY106 pKa = 10.46LARR109 pKa = 11.84YY110 pKa = 8.1MGGLMAKK117 pKa = 10.0RR118 pKa = 11.84FARR121 pKa = 11.84YY122 pKa = 8.87PEE124 pKa = 3.93LAAAEE129 pKa = 4.29VVMPVPLFPRR139 pKa = 11.84RR140 pKa = 11.84GRR142 pKa = 11.84KK143 pKa = 8.94RR144 pKa = 11.84GYY146 pKa = 9.28NQSEE150 pKa = 3.76LLARR154 pKa = 11.84FFAPQVGLPVQTKK167 pKa = 9.61ALVRR171 pKa = 11.84VRR173 pKa = 11.84NTVSQTKK180 pKa = 10.13LGRR183 pKa = 11.84AGRR186 pKa = 11.84LQNMAGAFACRR197 pKa = 11.84RR198 pKa = 11.84PEE200 pKa = 3.83WVKK203 pKa = 11.0GKK205 pKa = 8.03TVLLIDD211 pKa = 4.8DD212 pKa = 4.2VATTGATLEE221 pKa = 4.37GCAAALKK228 pKa = 10.27AAGARR233 pKa = 11.84RR234 pKa = 11.84VLAYY238 pKa = 9.2TFARR242 pKa = 11.84EE243 pKa = 3.86
Molecular weight: 26.91 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
459124 |
26 |
3027 |
329.1 |
36.51 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.044 ± 0.087 | 1.251 ± 0.029 |
4.839 ± 0.042 | 6.102 ± 0.055 |
4.544 ± 0.051 | 6.913 ± 0.061 |
1.769 ± 0.029 | 5.57 ± 0.059 |
6.804 ± 0.071 | 10.075 ± 0.069 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.415 ± 0.029 | 3.889 ± 0.041 |
4.544 ± 0.05 | 4.108 ± 0.055 |
4.839 ± 0.049 | 5.526 ± 0.048 |
5.39 ± 0.049 | 6.895 ± 0.053 |
1.093 ± 0.027 | 3.39 ± 0.037 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
