
Rosellinia necatrix megabirnavirus 2-W8
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Chrymotiviricetes; Ghabrivirales; Megabirnaviridae; Megabirnavirus; unclassified Megabirnavirus
Average proteome isoelectric point is 7.02
Get precalculated fractions of proteins
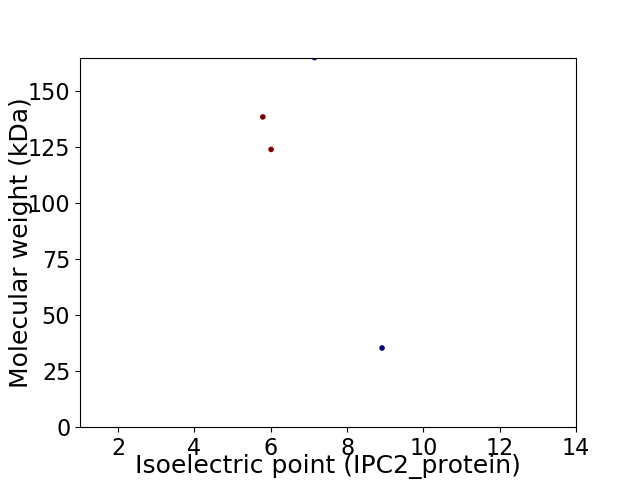
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0U5BZK0|A0A0U5BZK0_9VIRU Putative coat protein OS=Rosellinia necatrix megabirnavirus 2-W8 OX=1676267 PE=4 SV=1
MM1 pKa = 7.27VLNSYY6 pKa = 7.92NTATVPKK13 pKa = 10.24VVVNRR18 pKa = 11.84HH19 pKa = 5.31AFYY22 pKa = 11.14QNTKK26 pKa = 9.97AKK28 pKa = 10.2DD29 pKa = 3.4YY30 pKa = 10.16SFRR33 pKa = 11.84FARR36 pKa = 11.84EE37 pKa = 3.44DD38 pKa = 3.58DD39 pKa = 3.81AVNTLVAQLEE49 pKa = 4.64TGVVQHH55 pKa = 6.1QVGDD59 pKa = 3.69MNYY62 pKa = 9.51NAGGLGSFSTILSGKK77 pKa = 10.08KK78 pKa = 9.66KK79 pKa = 9.05VTHH82 pKa = 4.95QQIGAFIDD90 pKa = 3.85QTGHH94 pKa = 5.62GAEE97 pKa = 4.34GSIHH101 pKa = 5.42STSTVKK107 pKa = 10.6QYY109 pKa = 9.56GTGKK113 pKa = 9.48EE114 pKa = 3.95VAGAITDD121 pKa = 3.56VRR123 pKa = 11.84NMLGGVYY130 pKa = 9.77DD131 pKa = 3.94RR132 pKa = 11.84KK133 pKa = 10.64HH134 pKa = 6.22PGVHH138 pKa = 6.86KK139 pKa = 10.7LGTLAATYY147 pKa = 10.43TDD149 pKa = 4.54DD150 pKa = 5.53APDD153 pKa = 3.63GLTPIFEE160 pKa = 4.28RR161 pKa = 11.84LVRR164 pKa = 11.84GAMHH168 pKa = 5.37TTAYY172 pKa = 9.63NAHH175 pKa = 6.68RR176 pKa = 11.84QPAPVAGVAPADD188 pKa = 3.64PMGYY192 pKa = 7.9TPKK195 pKa = 10.33QYY197 pKa = 10.51YY198 pKa = 9.28HH199 pKa = 7.42DD200 pKa = 3.95SRR202 pKa = 11.84RR203 pKa = 11.84IMPHH207 pKa = 5.44RR208 pKa = 11.84QVALVPQAANVMPHH222 pKa = 6.85ASLINNSAEE231 pKa = 4.07NILANMDD238 pKa = 3.66VNVTNEE244 pKa = 3.91LTRR247 pKa = 11.84TLLTISEE254 pKa = 4.46VPSFRR259 pKa = 11.84AAEE262 pKa = 3.95LRR264 pKa = 11.84DD265 pKa = 3.43IVAVLDD271 pKa = 3.91ACTKK275 pKa = 10.72ANLGFDD281 pKa = 3.4VHH283 pKa = 7.51ADD285 pKa = 3.28QMNEE289 pKa = 3.42HH290 pKa = 6.29STYY293 pKa = 7.83THH295 pKa = 5.95GVSRR299 pKa = 11.84VHH301 pKa = 7.29DD302 pKa = 3.96FLILDD307 pKa = 3.47MCNFYY312 pKa = 11.01SRR314 pKa = 11.84CFTDD318 pKa = 3.83EE319 pKa = 3.85AAANDD324 pKa = 3.77NGFEE328 pKa = 4.17NLNDD332 pKa = 4.1PAPNLIPLPANNGLQPGVALWFGFAGNAANRR363 pKa = 11.84TGVCNPRR370 pKa = 11.84SWMNAIAFLLEE381 pKa = 4.13LEE383 pKa = 4.78GGRR386 pKa = 11.84EE387 pKa = 3.85ACAIAFNNVVEE398 pKa = 4.19QTTRR402 pKa = 11.84FFGPNVTMLASPPVVRR418 pKa = 11.84ADD420 pKa = 3.47PDD422 pKa = 4.04TFGSAHH428 pKa = 5.9RR429 pKa = 11.84VLRR432 pKa = 11.84QRR434 pKa = 11.84LGWMYY439 pKa = 10.76CNIIRR444 pKa = 11.84TPNSAGDD451 pKa = 3.17AWLDD455 pKa = 3.51KK456 pKa = 10.84QHH458 pKa = 6.14NQWPTHH464 pKa = 5.91PKK466 pKa = 9.7LGRR469 pKa = 11.84YY470 pKa = 8.85DD471 pKa = 3.81DD472 pKa = 3.93RR473 pKa = 11.84AAAAGAGAANAATIDD488 pKa = 3.42QTLARR493 pKa = 11.84NLRR496 pKa = 11.84NEE498 pKa = 4.3LFNNAAAAAPGALEE512 pKa = 3.71NARR515 pKa = 11.84FDD517 pKa = 4.57FVWADD522 pKa = 3.51VAHH525 pKa = 6.62NGPGQAMLEE534 pKa = 4.13NMSGGVDD541 pKa = 3.65PLNDD545 pKa = 3.23QLVRR549 pKa = 11.84RR550 pKa = 11.84WGDD553 pKa = 3.26YY554 pKa = 10.86VPIFGTGVAAANPGEE569 pKa = 4.5QNTAPPLGPLTITEE583 pKa = 4.25VDD585 pKa = 3.62NFWWRR590 pKa = 11.84FFLNVSLEE598 pKa = 3.99SDD600 pKa = 3.5EE601 pKa = 4.38VVEE604 pKa = 4.99DD605 pKa = 4.58FLRR608 pKa = 11.84LMPSSLLSEE617 pKa = 4.05LLIWLTDD624 pKa = 3.41SEE626 pKa = 4.83LRR628 pKa = 11.84GGDD631 pKa = 3.26AFNVRR636 pKa = 11.84WEE638 pKa = 4.38VPAALQVVDD647 pKa = 4.33GRR649 pKa = 11.84RR650 pKa = 11.84MTLPIPGNKK659 pKa = 7.33QHH661 pKa = 6.3YY662 pKa = 9.52ANPAHH667 pKa = 6.16MRR669 pKa = 11.84NFAWVNRR676 pKa = 11.84EE677 pKa = 3.73EE678 pKa = 3.79VHH680 pKa = 6.02YY681 pKa = 10.87ASEE684 pKa = 4.42FVGNICVPAYY694 pKa = 9.51TGLAAFWGYY703 pKa = 11.32VKK705 pKa = 9.83DD706 pKa = 3.7TRR708 pKa = 11.84TGRR711 pKa = 11.84VTEE714 pKa = 4.24DD715 pKa = 3.74DD716 pKa = 3.42EE717 pKa = 5.08RR718 pKa = 11.84LAARR722 pKa = 11.84FRR724 pKa = 11.84VMTSDD729 pKa = 3.73KK730 pKa = 11.07LMSLAFALGGQMRR743 pKa = 11.84CAADD747 pKa = 3.4AMAMEE752 pKa = 5.3LSLCRR757 pKa = 11.84STIARR762 pKa = 11.84MQGHH766 pKa = 7.39ILTDD770 pKa = 3.58NAALDD775 pKa = 4.09AEE777 pKa = 4.89LVDD780 pKa = 3.7TDD782 pKa = 4.83VINAQQSMKK791 pKa = 10.52GYY793 pKa = 10.57RR794 pKa = 11.84NTCLAEE800 pKa = 3.97YY801 pKa = 10.54DD802 pKa = 3.89SLAVGMGIYY811 pKa = 10.6TMMGTYY817 pKa = 9.76SRR819 pKa = 11.84YY820 pKa = 9.27VEE822 pKa = 4.0CDD824 pKa = 2.87VRR826 pKa = 11.84FNGEE830 pKa = 3.56TWNLRR835 pKa = 11.84RR836 pKa = 11.84NRR838 pKa = 11.84QTAFAAYY845 pKa = 9.22RR846 pKa = 11.84CLHH849 pKa = 6.5PLMFEE854 pKa = 4.18YY855 pKa = 10.58FCPGANMGGGMLAGQTQVEE874 pKa = 4.46RR875 pKa = 11.84VEE877 pKa = 4.42SGCVKK882 pKa = 9.67TFPALFKK889 pKa = 10.96PEE891 pKa = 4.61DD892 pKa = 3.82DD893 pKa = 4.79SIIDD897 pKa = 3.75LEE899 pKa = 4.52IYY901 pKa = 10.38AAAQQYY907 pKa = 10.88AIIAGYY913 pKa = 9.68HH914 pKa = 5.99IDD916 pKa = 3.38YY917 pKa = 10.62TMRR920 pKa = 11.84SLVKK924 pKa = 9.81RR925 pKa = 11.84TGDD928 pKa = 3.14GTGAQFYY935 pKa = 10.6EE936 pKa = 4.59LFNPYY941 pKa = 9.47LRR943 pKa = 11.84HH944 pKa = 6.38QIRR947 pKa = 11.84TSDD950 pKa = 4.15CGPGSPLLAVSPLSFYY966 pKa = 11.4VSTFNVDD973 pKa = 3.32LFEE976 pKa = 4.47QEE978 pKa = 4.44GSFKK982 pKa = 10.66VAKK985 pKa = 10.2LDD987 pKa = 3.91LAQQTFIHH995 pKa = 6.33RR996 pKa = 11.84TMGWFATTCGDD1007 pKa = 3.48AGRR1010 pKa = 11.84PNAFPHH1016 pKa = 5.66GTQRR1020 pKa = 11.84LANSDD1025 pKa = 2.95IGVNRR1030 pKa = 11.84RR1031 pKa = 11.84FGAAWGAHH1039 pKa = 4.32GTMKK1043 pKa = 9.08PTWGMATQRR1052 pKa = 11.84RR1053 pKa = 11.84RR1054 pKa = 11.84NRR1056 pKa = 11.84DD1057 pKa = 3.07DD1058 pKa = 5.18RR1059 pKa = 11.84FDD1061 pKa = 3.9LVATRR1066 pKa = 11.84VFEE1069 pKa = 4.3IQDD1072 pKa = 2.93ATGQVRR1078 pKa = 11.84RR1079 pKa = 11.84EE1080 pKa = 3.71YY1081 pKa = 10.9SVYY1084 pKa = 9.79TNQGFALVHH1093 pKa = 6.52APHH1096 pKa = 6.92GWDD1099 pKa = 2.44TWVQSNRR1106 pKa = 11.84GLIFTQSYY1114 pKa = 9.14GAAQHH1119 pKa = 5.13QQVDD1123 pKa = 4.07RR1124 pKa = 11.84SASGFKK1130 pKa = 10.15FCRR1133 pKa = 11.84NRR1135 pKa = 11.84LTFRR1139 pKa = 11.84MAAMSDD1145 pKa = 3.54DD1146 pKa = 4.57YY1147 pKa = 11.06EE1148 pKa = 4.5FTMHH1152 pKa = 7.41PLAKK1156 pKa = 10.19AEE1158 pKa = 4.0LMPVEE1163 pKa = 4.45KK1164 pKa = 9.98MEE1166 pKa = 3.95MTAVVTVGEE1175 pKa = 4.88GEE1177 pKa = 4.26TQSRR1181 pKa = 11.84VSTSRR1186 pKa = 11.84MGSALSDD1193 pKa = 3.3MGGKK1197 pKa = 10.06RR1198 pKa = 11.84GDD1200 pKa = 3.17GGLRR1204 pKa = 11.84PRR1206 pKa = 11.84GPRR1209 pKa = 11.84TNVTPVEE1216 pKa = 4.12KK1217 pKa = 9.87PIPGTVEE1224 pKa = 3.76HH1225 pKa = 6.94LEE1227 pKa = 4.28VPLTAGTPASQAATLVPEE1245 pKa = 4.11QGDD1248 pKa = 3.8RR1249 pKa = 11.84VSDD1252 pKa = 3.75PLVPKK1257 pKa = 10.65NN1258 pKa = 3.56
MM1 pKa = 7.27VLNSYY6 pKa = 7.92NTATVPKK13 pKa = 10.24VVVNRR18 pKa = 11.84HH19 pKa = 5.31AFYY22 pKa = 11.14QNTKK26 pKa = 9.97AKK28 pKa = 10.2DD29 pKa = 3.4YY30 pKa = 10.16SFRR33 pKa = 11.84FARR36 pKa = 11.84EE37 pKa = 3.44DD38 pKa = 3.58DD39 pKa = 3.81AVNTLVAQLEE49 pKa = 4.64TGVVQHH55 pKa = 6.1QVGDD59 pKa = 3.69MNYY62 pKa = 9.51NAGGLGSFSTILSGKK77 pKa = 10.08KK78 pKa = 9.66KK79 pKa = 9.05VTHH82 pKa = 4.95QQIGAFIDD90 pKa = 3.85QTGHH94 pKa = 5.62GAEE97 pKa = 4.34GSIHH101 pKa = 5.42STSTVKK107 pKa = 10.6QYY109 pKa = 9.56GTGKK113 pKa = 9.48EE114 pKa = 3.95VAGAITDD121 pKa = 3.56VRR123 pKa = 11.84NMLGGVYY130 pKa = 9.77DD131 pKa = 3.94RR132 pKa = 11.84KK133 pKa = 10.64HH134 pKa = 6.22PGVHH138 pKa = 6.86KK139 pKa = 10.7LGTLAATYY147 pKa = 10.43TDD149 pKa = 4.54DD150 pKa = 5.53APDD153 pKa = 3.63GLTPIFEE160 pKa = 4.28RR161 pKa = 11.84LVRR164 pKa = 11.84GAMHH168 pKa = 5.37TTAYY172 pKa = 9.63NAHH175 pKa = 6.68RR176 pKa = 11.84QPAPVAGVAPADD188 pKa = 3.64PMGYY192 pKa = 7.9TPKK195 pKa = 10.33QYY197 pKa = 10.51YY198 pKa = 9.28HH199 pKa = 7.42DD200 pKa = 3.95SRR202 pKa = 11.84RR203 pKa = 11.84IMPHH207 pKa = 5.44RR208 pKa = 11.84QVALVPQAANVMPHH222 pKa = 6.85ASLINNSAEE231 pKa = 4.07NILANMDD238 pKa = 3.66VNVTNEE244 pKa = 3.91LTRR247 pKa = 11.84TLLTISEE254 pKa = 4.46VPSFRR259 pKa = 11.84AAEE262 pKa = 3.95LRR264 pKa = 11.84DD265 pKa = 3.43IVAVLDD271 pKa = 3.91ACTKK275 pKa = 10.72ANLGFDD281 pKa = 3.4VHH283 pKa = 7.51ADD285 pKa = 3.28QMNEE289 pKa = 3.42HH290 pKa = 6.29STYY293 pKa = 7.83THH295 pKa = 5.95GVSRR299 pKa = 11.84VHH301 pKa = 7.29DD302 pKa = 3.96FLILDD307 pKa = 3.47MCNFYY312 pKa = 11.01SRR314 pKa = 11.84CFTDD318 pKa = 3.83EE319 pKa = 3.85AAANDD324 pKa = 3.77NGFEE328 pKa = 4.17NLNDD332 pKa = 4.1PAPNLIPLPANNGLQPGVALWFGFAGNAANRR363 pKa = 11.84TGVCNPRR370 pKa = 11.84SWMNAIAFLLEE381 pKa = 4.13LEE383 pKa = 4.78GGRR386 pKa = 11.84EE387 pKa = 3.85ACAIAFNNVVEE398 pKa = 4.19QTTRR402 pKa = 11.84FFGPNVTMLASPPVVRR418 pKa = 11.84ADD420 pKa = 3.47PDD422 pKa = 4.04TFGSAHH428 pKa = 5.9RR429 pKa = 11.84VLRR432 pKa = 11.84QRR434 pKa = 11.84LGWMYY439 pKa = 10.76CNIIRR444 pKa = 11.84TPNSAGDD451 pKa = 3.17AWLDD455 pKa = 3.51KK456 pKa = 10.84QHH458 pKa = 6.14NQWPTHH464 pKa = 5.91PKK466 pKa = 9.7LGRR469 pKa = 11.84YY470 pKa = 8.85DD471 pKa = 3.81DD472 pKa = 3.93RR473 pKa = 11.84AAAAGAGAANAATIDD488 pKa = 3.42QTLARR493 pKa = 11.84NLRR496 pKa = 11.84NEE498 pKa = 4.3LFNNAAAAAPGALEE512 pKa = 3.71NARR515 pKa = 11.84FDD517 pKa = 4.57FVWADD522 pKa = 3.51VAHH525 pKa = 6.62NGPGQAMLEE534 pKa = 4.13NMSGGVDD541 pKa = 3.65PLNDD545 pKa = 3.23QLVRR549 pKa = 11.84RR550 pKa = 11.84WGDD553 pKa = 3.26YY554 pKa = 10.86VPIFGTGVAAANPGEE569 pKa = 4.5QNTAPPLGPLTITEE583 pKa = 4.25VDD585 pKa = 3.62NFWWRR590 pKa = 11.84FFLNVSLEE598 pKa = 3.99SDD600 pKa = 3.5EE601 pKa = 4.38VVEE604 pKa = 4.99DD605 pKa = 4.58FLRR608 pKa = 11.84LMPSSLLSEE617 pKa = 4.05LLIWLTDD624 pKa = 3.41SEE626 pKa = 4.83LRR628 pKa = 11.84GGDD631 pKa = 3.26AFNVRR636 pKa = 11.84WEE638 pKa = 4.38VPAALQVVDD647 pKa = 4.33GRR649 pKa = 11.84RR650 pKa = 11.84MTLPIPGNKK659 pKa = 7.33QHH661 pKa = 6.3YY662 pKa = 9.52ANPAHH667 pKa = 6.16MRR669 pKa = 11.84NFAWVNRR676 pKa = 11.84EE677 pKa = 3.73EE678 pKa = 3.79VHH680 pKa = 6.02YY681 pKa = 10.87ASEE684 pKa = 4.42FVGNICVPAYY694 pKa = 9.51TGLAAFWGYY703 pKa = 11.32VKK705 pKa = 9.83DD706 pKa = 3.7TRR708 pKa = 11.84TGRR711 pKa = 11.84VTEE714 pKa = 4.24DD715 pKa = 3.74DD716 pKa = 3.42EE717 pKa = 5.08RR718 pKa = 11.84LAARR722 pKa = 11.84FRR724 pKa = 11.84VMTSDD729 pKa = 3.73KK730 pKa = 11.07LMSLAFALGGQMRR743 pKa = 11.84CAADD747 pKa = 3.4AMAMEE752 pKa = 5.3LSLCRR757 pKa = 11.84STIARR762 pKa = 11.84MQGHH766 pKa = 7.39ILTDD770 pKa = 3.58NAALDD775 pKa = 4.09AEE777 pKa = 4.89LVDD780 pKa = 3.7TDD782 pKa = 4.83VINAQQSMKK791 pKa = 10.52GYY793 pKa = 10.57RR794 pKa = 11.84NTCLAEE800 pKa = 3.97YY801 pKa = 10.54DD802 pKa = 3.89SLAVGMGIYY811 pKa = 10.6TMMGTYY817 pKa = 9.76SRR819 pKa = 11.84YY820 pKa = 9.27VEE822 pKa = 4.0CDD824 pKa = 2.87VRR826 pKa = 11.84FNGEE830 pKa = 3.56TWNLRR835 pKa = 11.84RR836 pKa = 11.84NRR838 pKa = 11.84QTAFAAYY845 pKa = 9.22RR846 pKa = 11.84CLHH849 pKa = 6.5PLMFEE854 pKa = 4.18YY855 pKa = 10.58FCPGANMGGGMLAGQTQVEE874 pKa = 4.46RR875 pKa = 11.84VEE877 pKa = 4.42SGCVKK882 pKa = 9.67TFPALFKK889 pKa = 10.96PEE891 pKa = 4.61DD892 pKa = 3.82DD893 pKa = 4.79SIIDD897 pKa = 3.75LEE899 pKa = 4.52IYY901 pKa = 10.38AAAQQYY907 pKa = 10.88AIIAGYY913 pKa = 9.68HH914 pKa = 5.99IDD916 pKa = 3.38YY917 pKa = 10.62TMRR920 pKa = 11.84SLVKK924 pKa = 9.81RR925 pKa = 11.84TGDD928 pKa = 3.14GTGAQFYY935 pKa = 10.6EE936 pKa = 4.59LFNPYY941 pKa = 9.47LRR943 pKa = 11.84HH944 pKa = 6.38QIRR947 pKa = 11.84TSDD950 pKa = 4.15CGPGSPLLAVSPLSFYY966 pKa = 11.4VSTFNVDD973 pKa = 3.32LFEE976 pKa = 4.47QEE978 pKa = 4.44GSFKK982 pKa = 10.66VAKK985 pKa = 10.2LDD987 pKa = 3.91LAQQTFIHH995 pKa = 6.33RR996 pKa = 11.84TMGWFATTCGDD1007 pKa = 3.48AGRR1010 pKa = 11.84PNAFPHH1016 pKa = 5.66GTQRR1020 pKa = 11.84LANSDD1025 pKa = 2.95IGVNRR1030 pKa = 11.84RR1031 pKa = 11.84FGAAWGAHH1039 pKa = 4.32GTMKK1043 pKa = 9.08PTWGMATQRR1052 pKa = 11.84RR1053 pKa = 11.84RR1054 pKa = 11.84NRR1056 pKa = 11.84DD1057 pKa = 3.07DD1058 pKa = 5.18RR1059 pKa = 11.84FDD1061 pKa = 3.9LVATRR1066 pKa = 11.84VFEE1069 pKa = 4.3IQDD1072 pKa = 2.93ATGQVRR1078 pKa = 11.84RR1079 pKa = 11.84EE1080 pKa = 3.71YY1081 pKa = 10.9SVYY1084 pKa = 9.79TNQGFALVHH1093 pKa = 6.52APHH1096 pKa = 6.92GWDD1099 pKa = 2.44TWVQSNRR1106 pKa = 11.84GLIFTQSYY1114 pKa = 9.14GAAQHH1119 pKa = 5.13QQVDD1123 pKa = 4.07RR1124 pKa = 11.84SASGFKK1130 pKa = 10.15FCRR1133 pKa = 11.84NRR1135 pKa = 11.84LTFRR1139 pKa = 11.84MAAMSDD1145 pKa = 3.54DD1146 pKa = 4.57YY1147 pKa = 11.06EE1148 pKa = 4.5FTMHH1152 pKa = 7.41PLAKK1156 pKa = 10.19AEE1158 pKa = 4.0LMPVEE1163 pKa = 4.45KK1164 pKa = 9.98MEE1166 pKa = 3.95MTAVVTVGEE1175 pKa = 4.88GEE1177 pKa = 4.26TQSRR1181 pKa = 11.84VSTSRR1186 pKa = 11.84MGSALSDD1193 pKa = 3.3MGGKK1197 pKa = 10.06RR1198 pKa = 11.84GDD1200 pKa = 3.17GGLRR1204 pKa = 11.84PRR1206 pKa = 11.84GPRR1209 pKa = 11.84TNVTPVEE1216 pKa = 4.12KK1217 pKa = 9.87PIPGTVEE1224 pKa = 3.76HH1225 pKa = 6.94LEE1227 pKa = 4.28VPLTAGTPASQAATLVPEE1245 pKa = 4.11QGDD1248 pKa = 3.8RR1249 pKa = 11.84VSDD1252 pKa = 3.75PLVPKK1257 pKa = 10.65NN1258 pKa = 3.56
Molecular weight: 138.39 kDa
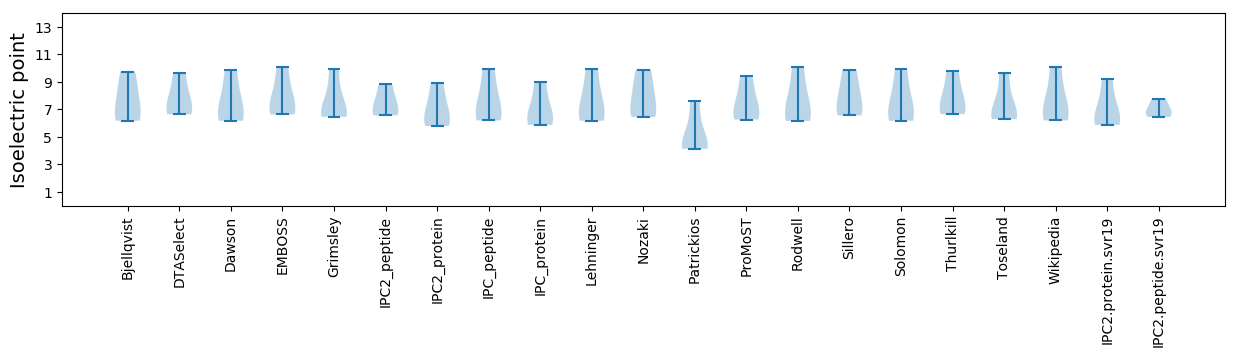
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0U5BZK0|A0A0U5BZK0_9VIRU Putative coat protein OS=Rosellinia necatrix megabirnavirus 2-W8 OX=1676267 PE=4 SV=1
MM1 pKa = 7.83ASVTKK6 pKa = 10.51KK7 pKa = 10.02LAKK10 pKa = 9.65VVCAIAAVRR19 pKa = 11.84QLHH22 pKa = 6.23RR23 pKa = 11.84MVDD26 pKa = 3.66LRR28 pKa = 11.84NVAMAARR35 pKa = 11.84CKK37 pKa = 9.68ATAEE41 pKa = 3.72WRR43 pKa = 11.84RR44 pKa = 11.84VLKK47 pKa = 10.79LVMCVMRR54 pKa = 11.84WQRR57 pKa = 11.84LVRR60 pKa = 11.84NSTSAASVTLSQLAEE75 pKa = 4.04QTTNAAGMGEE85 pKa = 4.28GDD87 pKa = 4.82VIWLAAIEE95 pKa = 4.2ALVSAVAVGPSVGAQXKK112 pKa = 9.17EE113 pKa = 4.03KK114 pKa = 10.68GVNQPGRR121 pKa = 11.84QLRR124 pKa = 11.84PVEE127 pKa = 4.4EE128 pKa = 4.34IGSLEE133 pKa = 4.05HH134 pKa = 5.69VAGVLRR140 pKa = 11.84SFLTWQRR147 pKa = 11.84YY148 pKa = 7.23RR149 pKa = 11.84RR150 pKa = 11.84IKK152 pKa = 9.7QLCFYY157 pKa = 8.3EE158 pKa = 4.16ARR160 pKa = 11.84RR161 pKa = 11.84IAAVRR166 pKa = 11.84NLRR169 pKa = 11.84IAVAARR175 pKa = 11.84DD176 pKa = 3.71EE177 pKa = 4.34AAATPGYY184 pKa = 9.41HH185 pKa = 3.59QTARR189 pKa = 11.84YY190 pKa = 7.64VHH192 pKa = 6.63SMLCKK197 pKa = 9.7RR198 pKa = 11.84AEE200 pKa = 4.3VPCTCGGLWVDD211 pKa = 3.59EE212 pKa = 4.89DD213 pKa = 4.35GAVLGEE219 pKa = 4.2AQAAAAFSSVQNQGTPRR236 pKa = 11.84FSPATHH242 pKa = 6.85TIVVALPVRR251 pKa = 11.84TEE253 pKa = 4.06SQRR256 pKa = 11.84SIAPNNQATAGDD268 pKa = 3.9RR269 pKa = 11.84QEE271 pKa = 5.34HH272 pKa = 5.63VGQLSMQEE280 pKa = 3.86LVAAYY285 pKa = 9.27IAEE288 pKa = 4.25HH289 pKa = 6.59RR290 pKa = 11.84SGVQAALRR298 pKa = 11.84MRR300 pKa = 11.84WQLPEE305 pKa = 4.44GVDD308 pKa = 3.27ANEE311 pKa = 4.08FVTAVAQRR319 pKa = 11.84LKK321 pKa = 11.01EE322 pKa = 4.03SDD324 pKa = 3.2QQ325 pKa = 3.44
MM1 pKa = 7.83ASVTKK6 pKa = 10.51KK7 pKa = 10.02LAKK10 pKa = 9.65VVCAIAAVRR19 pKa = 11.84QLHH22 pKa = 6.23RR23 pKa = 11.84MVDD26 pKa = 3.66LRR28 pKa = 11.84NVAMAARR35 pKa = 11.84CKK37 pKa = 9.68ATAEE41 pKa = 3.72WRR43 pKa = 11.84RR44 pKa = 11.84VLKK47 pKa = 10.79LVMCVMRR54 pKa = 11.84WQRR57 pKa = 11.84LVRR60 pKa = 11.84NSTSAASVTLSQLAEE75 pKa = 4.04QTTNAAGMGEE85 pKa = 4.28GDD87 pKa = 4.82VIWLAAIEE95 pKa = 4.2ALVSAVAVGPSVGAQXKK112 pKa = 9.17EE113 pKa = 4.03KK114 pKa = 10.68GVNQPGRR121 pKa = 11.84QLRR124 pKa = 11.84PVEE127 pKa = 4.4EE128 pKa = 4.34IGSLEE133 pKa = 4.05HH134 pKa = 5.69VAGVLRR140 pKa = 11.84SFLTWQRR147 pKa = 11.84YY148 pKa = 7.23RR149 pKa = 11.84RR150 pKa = 11.84IKK152 pKa = 9.7QLCFYY157 pKa = 8.3EE158 pKa = 4.16ARR160 pKa = 11.84RR161 pKa = 11.84IAAVRR166 pKa = 11.84NLRR169 pKa = 11.84IAVAARR175 pKa = 11.84DD176 pKa = 3.71EE177 pKa = 4.34AAATPGYY184 pKa = 9.41HH185 pKa = 3.59QTARR189 pKa = 11.84YY190 pKa = 7.64VHH192 pKa = 6.63SMLCKK197 pKa = 9.7RR198 pKa = 11.84AEE200 pKa = 4.3VPCTCGGLWVDD211 pKa = 3.59EE212 pKa = 4.89DD213 pKa = 4.35GAVLGEE219 pKa = 4.2AQAAAAFSSVQNQGTPRR236 pKa = 11.84FSPATHH242 pKa = 6.85TIVVALPVRR251 pKa = 11.84TEE253 pKa = 4.06SQRR256 pKa = 11.84SIAPNNQATAGDD268 pKa = 3.9RR269 pKa = 11.84QEE271 pKa = 5.34HH272 pKa = 5.63VGQLSMQEE280 pKa = 3.86LVAAYY285 pKa = 9.27IAEE288 pKa = 4.25HH289 pKa = 6.59RR290 pKa = 11.84SGVQAALRR298 pKa = 11.84MRR300 pKa = 11.84WQLPEE305 pKa = 4.44GVDD308 pKa = 3.27ANEE311 pKa = 4.08FVTAVAQRR319 pKa = 11.84LKK321 pKa = 11.01EE322 pKa = 4.03SDD324 pKa = 3.2QQ325 pKa = 3.44
Molecular weight: 35.33 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4221 |
325 |
1530 |
1055.3 |
115.61 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.888 ± 1.149 | 1.327 ± 0.126 |
5.33 ± 0.575 | 4.928 ± 0.215 |
3.435 ± 0.476 | 7.818 ± 0.477 |
2.464 ± 0.17 | 3.364 ± 0.107 |
2.227 ± 0.45 | 8.742 ± 0.613 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.175 ± 0.253 | 3.909 ± 0.628 |
4.454 ± 0.305 | 4.241 ± 0.382 |
7.865 ± 0.422 | 5.354 ± 0.769 |
6.207 ± 0.753 | 7.771 ± 0.612 |
1.943 ± 0.147 | 2.535 ± 0.398 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
