
Microviridae Bog9017_22
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 6.98
Get precalculated fractions of proteins
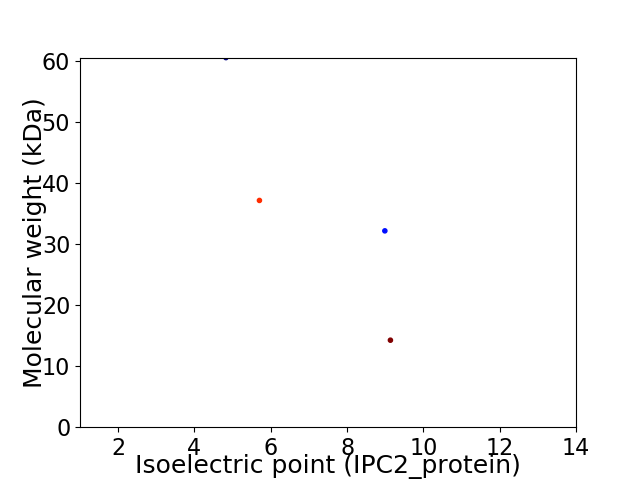
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0G2UK20|A0A0G2UK20_9VIRU DNA pilot protein VP2 OS=Microviridae Bog9017_22 OX=1655651 PE=4 SV=1
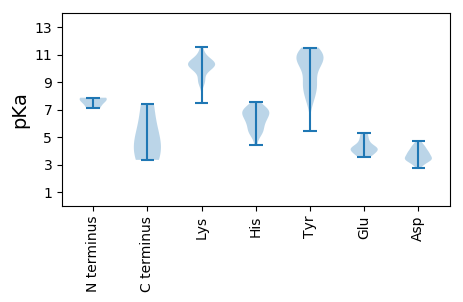
MM1 pKa = 7.08STQKK5 pKa = 10.36IIQTGKK11 pKa = 9.05VGLVSRR17 pKa = 11.84PNNSVPSNTFDD28 pKa = 3.87LSYY31 pKa = 11.11DD32 pKa = 3.33VKK34 pKa = 10.05TSFNIGEE41 pKa = 5.28LIPIGCWEE49 pKa = 4.74ALPGDD54 pKa = 4.05RR55 pKa = 11.84FNCQSEE61 pKa = 4.2ALVRR65 pKa = 11.84FAPMLAPIMQRR76 pKa = 11.84LNLRR80 pKa = 11.84LEE82 pKa = 4.22YY83 pKa = 10.6FFVPNRR89 pKa = 11.84ILWSDD94 pKa = 3.14WEE96 pKa = 4.55DD97 pKa = 4.2FISPSTEE104 pKa = 3.9GEE106 pKa = 4.07ASPSMAAFSDD116 pKa = 3.91DD117 pKa = 3.95FQVTPYY123 pKa = 10.5SVSGYY128 pKa = 10.57LGLPLTGPDD137 pKa = 3.73LVISKK142 pKa = 10.87DD143 pKa = 3.54DD144 pKa = 3.92VNCLRR149 pKa = 11.84HH150 pKa = 5.4AGYY153 pKa = 10.35RR154 pKa = 11.84RR155 pKa = 11.84IYY157 pKa = 9.05EE158 pKa = 3.55DD159 pKa = 3.07WYY161 pKa = 10.94RR162 pKa = 11.84DD163 pKa = 3.31EE164 pKa = 5.26NYY166 pKa = 11.13DD167 pKa = 3.4MSGGNTSLSSGFTNQFDD184 pKa = 3.84IYY186 pKa = 11.32DD187 pKa = 4.12HH188 pKa = 6.8IFTRR192 pKa = 11.84NLDD195 pKa = 3.22RR196 pKa = 11.84DD197 pKa = 4.38YY198 pKa = 10.66FTSSYY203 pKa = 10.46PWPQKK208 pKa = 10.64GADD211 pKa = 3.38VLIPVTIDD219 pKa = 2.9QTPQVVPVSRR229 pKa = 11.84TSQGVYY235 pKa = 8.59TVFEE239 pKa = 4.25PVDD242 pKa = 3.67NADD245 pKa = 3.52HH246 pKa = 7.01SGGAVDD252 pKa = 4.34WVPDD256 pKa = 4.32PEE258 pKa = 4.81PSSNTGYY265 pKa = 8.77LTAGSTPLSIDD276 pKa = 3.41PGSSGFQVSIPADD289 pKa = 3.43SSTVGTINDD298 pKa = 3.96LRR300 pKa = 11.84TAAAVQRR307 pKa = 11.84WLEE310 pKa = 3.57ADD312 pKa = 3.11IYY314 pKa = 7.5GTRR317 pKa = 11.84YY318 pKa = 9.45IEE320 pKa = 5.0LIEE323 pKa = 4.01NHH325 pKa = 6.25FATTVEE331 pKa = 4.02DD332 pKa = 4.25FRR334 pKa = 11.84LQRR337 pKa = 11.84AEE339 pKa = 3.95FLGMTLEE346 pKa = 5.26PIQISEE352 pKa = 4.26VLQTSEE358 pKa = 4.53TATSPQGNMSGHH370 pKa = 6.24GLAVHH375 pKa = 6.77KK376 pKa = 10.68GEE378 pKa = 4.99HH379 pKa = 5.91YY380 pKa = 10.57NYY382 pKa = 8.8HH383 pKa = 6.27CKK385 pKa = 10.05EE386 pKa = 3.92HH387 pKa = 5.43GHH389 pKa = 6.28FYY391 pKa = 10.85VIASVLPTNAYY402 pKa = 7.72YY403 pKa = 10.42QGVARR408 pKa = 11.84GFSRR412 pKa = 11.84FDD414 pKa = 3.36RR415 pKa = 11.84LDD417 pKa = 3.54FAWPEE422 pKa = 3.68LANIGFQEE430 pKa = 4.35LLNKK434 pKa = 9.94EE435 pKa = 4.16LFYY438 pKa = 11.29DD439 pKa = 4.71VSDD442 pKa = 4.61DD443 pKa = 4.27LNGDD447 pKa = 3.39VFGYY451 pKa = 8.88VPRR454 pKa = 11.84YY455 pKa = 8.93NEE457 pKa = 3.65YY458 pKa = 8.67RR459 pKa = 11.84TMYY462 pKa = 10.07SQVTGEE468 pKa = 4.13FATNLQYY475 pKa = 11.06WHH477 pKa = 7.13MGRR480 pKa = 11.84DD481 pKa = 3.61FASRR485 pKa = 11.84PTHH488 pKa = 5.22TEE490 pKa = 3.53PFNTTDD496 pKa = 3.33PSTVDD501 pKa = 2.77RR502 pKa = 11.84VFAVVEE508 pKa = 4.39STVSHH513 pKa = 6.62LYY515 pKa = 9.12VHH517 pKa = 7.31AFHH520 pKa = 6.94KK521 pKa = 10.79CKK523 pKa = 10.59VNRR526 pKa = 11.84KK527 pKa = 8.58LPRR530 pKa = 11.84FSVPYY535 pKa = 9.92LHH537 pKa = 7.4
MM1 pKa = 7.08STQKK5 pKa = 10.36IIQTGKK11 pKa = 9.05VGLVSRR17 pKa = 11.84PNNSVPSNTFDD28 pKa = 3.87LSYY31 pKa = 11.11DD32 pKa = 3.33VKK34 pKa = 10.05TSFNIGEE41 pKa = 5.28LIPIGCWEE49 pKa = 4.74ALPGDD54 pKa = 4.05RR55 pKa = 11.84FNCQSEE61 pKa = 4.2ALVRR65 pKa = 11.84FAPMLAPIMQRR76 pKa = 11.84LNLRR80 pKa = 11.84LEE82 pKa = 4.22YY83 pKa = 10.6FFVPNRR89 pKa = 11.84ILWSDD94 pKa = 3.14WEE96 pKa = 4.55DD97 pKa = 4.2FISPSTEE104 pKa = 3.9GEE106 pKa = 4.07ASPSMAAFSDD116 pKa = 3.91DD117 pKa = 3.95FQVTPYY123 pKa = 10.5SVSGYY128 pKa = 10.57LGLPLTGPDD137 pKa = 3.73LVISKK142 pKa = 10.87DD143 pKa = 3.54DD144 pKa = 3.92VNCLRR149 pKa = 11.84HH150 pKa = 5.4AGYY153 pKa = 10.35RR154 pKa = 11.84RR155 pKa = 11.84IYY157 pKa = 9.05EE158 pKa = 3.55DD159 pKa = 3.07WYY161 pKa = 10.94RR162 pKa = 11.84DD163 pKa = 3.31EE164 pKa = 5.26NYY166 pKa = 11.13DD167 pKa = 3.4MSGGNTSLSSGFTNQFDD184 pKa = 3.84IYY186 pKa = 11.32DD187 pKa = 4.12HH188 pKa = 6.8IFTRR192 pKa = 11.84NLDD195 pKa = 3.22RR196 pKa = 11.84DD197 pKa = 4.38YY198 pKa = 10.66FTSSYY203 pKa = 10.46PWPQKK208 pKa = 10.64GADD211 pKa = 3.38VLIPVTIDD219 pKa = 2.9QTPQVVPVSRR229 pKa = 11.84TSQGVYY235 pKa = 8.59TVFEE239 pKa = 4.25PVDD242 pKa = 3.67NADD245 pKa = 3.52HH246 pKa = 7.01SGGAVDD252 pKa = 4.34WVPDD256 pKa = 4.32PEE258 pKa = 4.81PSSNTGYY265 pKa = 8.77LTAGSTPLSIDD276 pKa = 3.41PGSSGFQVSIPADD289 pKa = 3.43SSTVGTINDD298 pKa = 3.96LRR300 pKa = 11.84TAAAVQRR307 pKa = 11.84WLEE310 pKa = 3.57ADD312 pKa = 3.11IYY314 pKa = 7.5GTRR317 pKa = 11.84YY318 pKa = 9.45IEE320 pKa = 5.0LIEE323 pKa = 4.01NHH325 pKa = 6.25FATTVEE331 pKa = 4.02DD332 pKa = 4.25FRR334 pKa = 11.84LQRR337 pKa = 11.84AEE339 pKa = 3.95FLGMTLEE346 pKa = 5.26PIQISEE352 pKa = 4.26VLQTSEE358 pKa = 4.53TATSPQGNMSGHH370 pKa = 6.24GLAVHH375 pKa = 6.77KK376 pKa = 10.68GEE378 pKa = 4.99HH379 pKa = 5.91YY380 pKa = 10.57NYY382 pKa = 8.8HH383 pKa = 6.27CKK385 pKa = 10.05EE386 pKa = 3.92HH387 pKa = 5.43GHH389 pKa = 6.28FYY391 pKa = 10.85VIASVLPTNAYY402 pKa = 7.72YY403 pKa = 10.42QGVARR408 pKa = 11.84GFSRR412 pKa = 11.84FDD414 pKa = 3.36RR415 pKa = 11.84LDD417 pKa = 3.54FAWPEE422 pKa = 3.68LANIGFQEE430 pKa = 4.35LLNKK434 pKa = 9.94EE435 pKa = 4.16LFYY438 pKa = 11.29DD439 pKa = 4.71VSDD442 pKa = 4.61DD443 pKa = 4.27LNGDD447 pKa = 3.39VFGYY451 pKa = 8.88VPRR454 pKa = 11.84YY455 pKa = 8.93NEE457 pKa = 3.65YY458 pKa = 8.67RR459 pKa = 11.84TMYY462 pKa = 10.07SQVTGEE468 pKa = 4.13FATNLQYY475 pKa = 11.06WHH477 pKa = 7.13MGRR480 pKa = 11.84DD481 pKa = 3.61FASRR485 pKa = 11.84PTHH488 pKa = 5.22TEE490 pKa = 3.53PFNTTDD496 pKa = 3.33PSTVDD501 pKa = 2.77RR502 pKa = 11.84VFAVVEE508 pKa = 4.39STVSHH513 pKa = 6.62LYY515 pKa = 9.12VHH517 pKa = 7.31AFHH520 pKa = 6.94KK521 pKa = 10.79CKK523 pKa = 10.59VNRR526 pKa = 11.84KK527 pKa = 8.58LPRR530 pKa = 11.84FSVPYY535 pKa = 9.92LHH537 pKa = 7.4
Molecular weight: 60.52 kDa
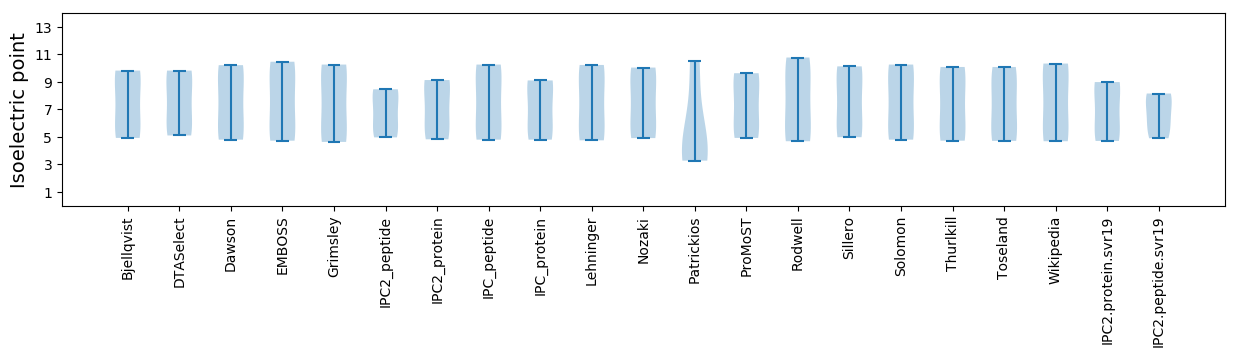
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0G2UMD9|A0A0G2UMD9_9VIRU Uncharacterized protein OS=Microviridae Bog9017_22 OX=1655651 PE=4 SV=1
MM1 pKa = 7.54NFTFQNVANWRR12 pKa = 11.84LLLNRR17 pKa = 11.84RR18 pKa = 11.84QTWPPSPVQPNLALSIKK35 pKa = 10.16DD36 pKa = 2.98IYY38 pKa = 11.39SRR40 pKa = 11.84FLRR43 pKa = 11.84RR44 pKa = 11.84QPLPDD49 pKa = 3.42VYY51 pKa = 11.17DD52 pKa = 4.22GFSSPDD58 pKa = 3.57DD59 pKa = 4.35LSDD62 pKa = 3.74FDD64 pKa = 4.62NLSKK68 pKa = 10.08IDD70 pKa = 3.27KK71 pKa = 10.14AAAAKK76 pKa = 10.11HH77 pKa = 4.39YY78 pKa = 9.65AKK80 pKa = 10.31RR81 pKa = 11.84VATLDD86 pKa = 3.28SDD88 pKa = 3.94LKK90 pKa = 11.03EE91 pKa = 3.88RR92 pKa = 11.84SKK94 pKa = 11.55KK95 pKa = 10.23NAFEE99 pKa = 3.82QAAARR104 pKa = 11.84HH105 pKa = 5.66RR106 pKa = 11.84KK107 pKa = 8.94SVADD111 pKa = 3.92EE112 pKa = 4.17VAKK115 pKa = 10.65EE116 pKa = 3.75LAKK119 pKa = 10.33TKK121 pKa = 10.08TVVSS125 pKa = 4.11
MM1 pKa = 7.54NFTFQNVANWRR12 pKa = 11.84LLLNRR17 pKa = 11.84RR18 pKa = 11.84QTWPPSPVQPNLALSIKK35 pKa = 10.16DD36 pKa = 2.98IYY38 pKa = 11.39SRR40 pKa = 11.84FLRR43 pKa = 11.84RR44 pKa = 11.84QPLPDD49 pKa = 3.42VYY51 pKa = 11.17DD52 pKa = 4.22GFSSPDD58 pKa = 3.57DD59 pKa = 4.35LSDD62 pKa = 3.74FDD64 pKa = 4.62NLSKK68 pKa = 10.08IDD70 pKa = 3.27KK71 pKa = 10.14AAAAKK76 pKa = 10.11HH77 pKa = 4.39YY78 pKa = 9.65AKK80 pKa = 10.31RR81 pKa = 11.84VATLDD86 pKa = 3.28SDD88 pKa = 3.94LKK90 pKa = 11.03EE91 pKa = 3.88RR92 pKa = 11.84SKK94 pKa = 11.55KK95 pKa = 10.23NAFEE99 pKa = 3.82QAAARR104 pKa = 11.84HH105 pKa = 5.66RR106 pKa = 11.84KK107 pKa = 8.94SVADD111 pKa = 3.92EE112 pKa = 4.17VAKK115 pKa = 10.65EE116 pKa = 3.75LAKK119 pKa = 10.33TKK121 pKa = 10.08TVVSS125 pKa = 4.11
Molecular weight: 14.25 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1291 |
125 |
537 |
322.8 |
36.01 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.985 ± 2.034 | 1.084 ± 0.5 |
6.197 ± 0.632 | 3.796 ± 0.689 |
4.493 ± 0.887 | 5.887 ± 0.955 |
2.479 ± 0.899 | 4.105 ± 0.319 |
4.183 ± 1.254 | 8.753 ± 0.86 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.169 ± 0.586 | 5.19 ± 0.814 |
5.422 ± 0.472 | 5.345 ± 1.207 |
5.422 ± 0.994 | 8.366 ± 0.223 |
5.577 ± 0.729 | 6.739 ± 0.634 |
1.162 ± 0.281 | 4.648 ± 1.088 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
