
Flexilinea flocculi
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Chloroflexi; Anaerolineae; Anaerolineales; Anaerolineaceae; Flexilinea
Average proteome isoelectric point is 6.21
Get precalculated fractions of proteins
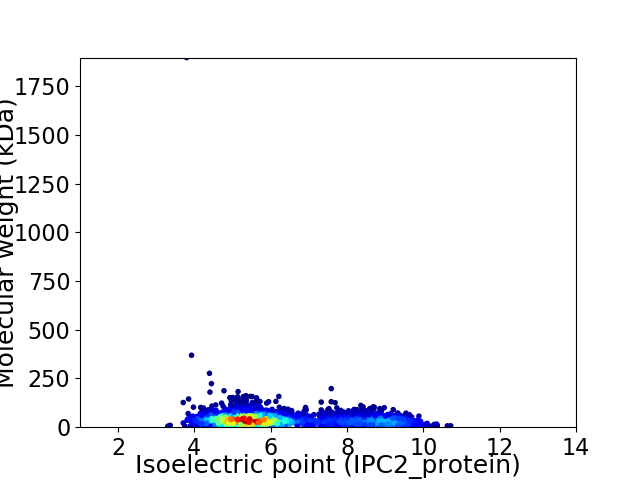
Virtual 2D-PAGE plot for 2774 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0K8PBE2|A0A0K8PBE2_9CHLR Cysteine synthase OS=Flexilinea flocculi OX=1678840 GN=ATC1_12517 PE=4 SV=1
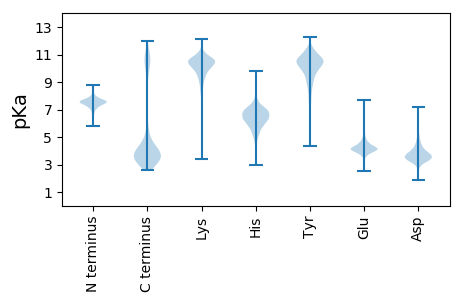
MM1 pKa = 7.54LWLFSDD7 pKa = 4.15IVQDD11 pKa = 4.54NIDD14 pKa = 4.27DD15 pKa = 3.92KK16 pKa = 11.22HH17 pKa = 7.15VKK19 pKa = 9.13YY20 pKa = 10.51YY21 pKa = 10.88YY22 pKa = 10.34YY23 pKa = 11.27GSTEE27 pKa = 3.77PDD29 pKa = 3.13NPEE32 pKa = 4.2NLDD35 pKa = 3.61PDD37 pKa = 3.56SWIEE41 pKa = 3.78TTYY44 pKa = 10.69YY45 pKa = 10.86GPISITITNDD55 pKa = 2.76VTNSQITFTNGASYY69 pKa = 10.28TPVSPTAGTTNQPIGGFSLQADD91 pKa = 4.55FPGASLTSVTISTTGTRR108 pKa = 11.84SGITNLKK115 pKa = 9.53LWSSTDD121 pKa = 3.01ATFNASSDD129 pKa = 3.63TLLNSQSDD137 pKa = 3.9NASVTFSGFSSTISTAGSYY156 pKa = 10.58YY157 pKa = 10.48FVTADD162 pKa = 4.18LSTSASGSIALTIGSVSNLTFSGGAIASGFTNAPLSSGTITIILIRR208 pKa = 11.84EE209 pKa = 3.9MNITGNGVTILNGDD223 pKa = 4.12TSPAASDD230 pKa = 3.37HH231 pKa = 6.37TDD233 pKa = 3.54FGSVDD238 pKa = 3.23IASGSVVHH246 pKa = 6.71TFTIQNNGAEE256 pKa = 4.42ALSLTGSSPFVAISGATADD275 pKa = 5.18FSLTQNPSSSIAGNSSTTFEE295 pKa = 4.03ITFNPASTGLKK306 pKa = 9.8SAVISIANDD315 pKa = 3.7DD316 pKa = 3.97GDD318 pKa = 4.18EE319 pKa = 4.1NPYY322 pKa = 9.89TFTIQGTGTAVPNMSVSGNAMQITNGDD349 pKa = 3.81IEE351 pKa = 4.5PSEE354 pKa = 5.06SDD356 pKa = 3.28DD357 pKa = 3.64TDD359 pKa = 3.78FGSVNIAGDD368 pKa = 4.27PIEE371 pKa = 4.35HH372 pKa = 6.78TFTIANNGSASLTLTGSSPYY392 pKa = 10.67VSLTGDD398 pKa = 3.56TEE400 pKa = 5.47DD401 pKa = 3.76FTLTQLPASNIAAGGGTTTFKK422 pKa = 9.92ITFKK426 pKa = 9.92PTATGNRR433 pKa = 11.84SATVSIANDD442 pKa = 3.89DD443 pKa = 3.92DD444 pKa = 4.34SKK446 pKa = 11.83NPFNFSIQGSGISVPTVTTAAASSVTKK473 pKa = 9.93TVATLGGNVTSDD485 pKa = 3.02GGSPITEE492 pKa = 4.01TGVVYY497 pKa = 10.61SSSNATPTIGGMDD510 pKa = 4.19VIQNANIDD518 pKa = 3.69LSGDD522 pKa = 3.44FSEE525 pKa = 5.52SISPLSSATHH535 pKa = 6.38YY536 pKa = 10.43YY537 pKa = 8.74FQAYY541 pKa = 6.59ATNAAGTSYY550 pKa = 11.18GGVLEE555 pKa = 4.38FTTLTSVVSILRR567 pKa = 11.84ADD569 pKa = 4.25PDD571 pKa = 3.71DD572 pKa = 3.95TNAASVRR579 pKa = 11.84WNVVFEE585 pKa = 4.42NNVTGLSASNFTLVSFGISGAAITGVSGSGTNWIVTASTGSGTGSLRR632 pKa = 11.84LGLVNDD638 pKa = 4.04NGLTPKK644 pKa = 10.56LSNPTLGGDD653 pKa = 3.91AYY655 pKa = 9.86TLDD658 pKa = 3.94LVSPSVTDD666 pKa = 3.32VDD668 pKa = 3.63TGKK671 pKa = 9.85PDD673 pKa = 3.05GTYY676 pKa = 10.99GVGSSIGIVIMFDD689 pKa = 3.09EE690 pKa = 4.4SHH692 pKa = 6.55NVDD695 pKa = 3.44GTPQLTLKK703 pKa = 8.88MDD705 pKa = 3.41SMNRR709 pKa = 11.84VIDD712 pKa = 4.74CISNEE717 pKa = 4.47HH718 pKa = 6.38IRR720 pKa = 11.84LICTYY725 pKa = 8.99TVQAGDD731 pKa = 3.56NTSDD735 pKa = 3.56LDD737 pKa = 4.12YY738 pKa = 11.62AGMTALSTNGGTIRR752 pKa = 11.84DD753 pKa = 3.79YY754 pKa = 11.22HH755 pKa = 7.8GNDD758 pKa = 4.57AILNLPWNAGTGSLAGNNAIVIDD781 pKa = 4.09TTPPEE786 pKa = 4.15ITIHH790 pKa = 6.26HH791 pKa = 7.19PGSSAAVSKK800 pKa = 9.77TITATTSDD808 pKa = 3.7GTIKK812 pKa = 10.23MSNTRR817 pKa = 11.84GTTCDD822 pKa = 2.97GSLIFTAYY830 pKa = 10.49ASQTFNAEE838 pKa = 3.55TDD840 pKa = 3.02NGIRR844 pKa = 11.84VCYY847 pKa = 9.75QAVDD851 pKa = 3.68TVNNISYY858 pKa = 10.4KK859 pKa = 10.63LSDD862 pKa = 5.22PISGIDD868 pKa = 3.3TTKK871 pKa = 9.28PTTSIIAKK879 pKa = 9.42PGNPDD884 pKa = 2.97NSASPSFTFSGEE896 pKa = 4.22DD897 pKa = 3.4PDD899 pKa = 4.5GTDD902 pKa = 3.65GSDD905 pKa = 3.15GSGIAGFEE913 pKa = 4.16CSLDD917 pKa = 3.6GEE919 pKa = 4.76PFSSCSSPKK928 pKa = 9.76TYY930 pKa = 10.92SSLGDD935 pKa = 3.6GNHH938 pKa = 5.92TFQVRR943 pKa = 11.84SVDD946 pKa = 3.45AAGNTDD952 pKa = 4.84DD953 pKa = 5.56SPEE956 pKa = 4.23SYY958 pKa = 8.6TWQIDD963 pKa = 3.37TSLPEE968 pKa = 4.05TTILSSPANPTASTDD983 pKa = 3.24AVFEE987 pKa = 4.27FEE989 pKa = 4.36GADD992 pKa = 3.43PGGSGVSGFEE1002 pKa = 4.05CSLDD1006 pKa = 3.55GEE1008 pKa = 4.6PFSDD1012 pKa = 3.81CSSPKK1017 pKa = 10.22SYY1019 pKa = 11.33SSLSDD1024 pKa = 3.39GNHH1027 pKa = 5.58TFLVRR1032 pKa = 11.84SVDD1035 pKa = 3.49AVGNADD1041 pKa = 3.55SSPAEE1046 pKa = 4.22FNWTVDD1052 pKa = 3.32TTPPEE1057 pKa = 4.07IAIHH1061 pKa = 6.17HH1062 pKa = 6.83PGSSAAVSKK1071 pKa = 9.77TITATTSDD1079 pKa = 3.7GTIKK1083 pKa = 10.23MSNTRR1088 pKa = 11.84GTTCDD1093 pKa = 3.06GSLTFTVYY1101 pKa = 10.91ASQTFNAEE1109 pKa = 3.55TDD1111 pKa = 3.02NGIRR1115 pKa = 11.84VCYY1118 pKa = 9.6QAVDD1122 pKa = 3.4AVNNISYY1129 pKa = 10.53KK1130 pKa = 10.7LSDD1133 pKa = 5.22PISGIDD1139 pKa = 3.3TTKK1142 pKa = 9.28PTTSIIAKK1150 pKa = 9.42PGNPDD1155 pKa = 2.97NSASPSFTFSGEE1167 pKa = 4.2DD1168 pKa = 3.38PDD1170 pKa = 5.4GADD1173 pKa = 3.81GSDD1176 pKa = 3.18GSGIAGFEE1184 pKa = 4.16CSLDD1188 pKa = 3.6GEE1190 pKa = 4.76PFSSCSSPKK1199 pKa = 9.76TYY1201 pKa = 10.92SSLGDD1206 pKa = 3.6GNHH1209 pKa = 5.92TFQVRR1214 pKa = 11.84SVDD1217 pKa = 3.45AAGNTDD1223 pKa = 4.84DD1224 pKa = 5.56SPEE1227 pKa = 4.23SYY1229 pKa = 8.6TWQIDD1234 pKa = 3.37TSLPEE1239 pKa = 4.05TTILSSPANLTTSTDD1254 pKa = 3.13AVFEE1258 pKa = 4.26FEE1260 pKa = 4.77GDD1262 pKa = 3.68DD1263 pKa = 3.68PGGSGIEE1270 pKa = 4.09NLEE1273 pKa = 4.11CMLDD1277 pKa = 3.26SSTFSVCTSPKK1288 pKa = 9.96EE1289 pKa = 4.09YY1290 pKa = 10.57NALPDD1295 pKa = 3.93GSHH1298 pKa = 6.17NFQVRR1303 pKa = 11.84AVDD1306 pKa = 3.32TAGNVDD1312 pKa = 4.29PSPAEE1317 pKa = 3.99INWTIDD1323 pKa = 3.03TTAPTVLISSTEE1335 pKa = 3.89SDD1337 pKa = 3.6PTNVNPIPITVQFSEE1352 pKa = 4.42PVTGFEE1358 pKa = 4.6EE1359 pKa = 4.48SDD1361 pKa = 2.95IMYY1364 pKa = 9.16HH1365 pKa = 5.71QRR1367 pKa = 11.84LPRR1370 pKa = 11.84SGQFHH1375 pKa = 6.71VFLQFDD1381 pKa = 3.88PEE1383 pKa = 4.06QRR1385 pKa = 11.84RR1386 pKa = 11.84DD1387 pKa = 3.17SYY1389 pKa = 11.32PRR1391 pKa = 11.84YY1392 pKa = 9.81SSSCRR1397 pKa = 11.84RR1398 pKa = 3.43
MM1 pKa = 7.54LWLFSDD7 pKa = 4.15IVQDD11 pKa = 4.54NIDD14 pKa = 4.27DD15 pKa = 3.92KK16 pKa = 11.22HH17 pKa = 7.15VKK19 pKa = 9.13YY20 pKa = 10.51YY21 pKa = 10.88YY22 pKa = 10.34YY23 pKa = 11.27GSTEE27 pKa = 3.77PDD29 pKa = 3.13NPEE32 pKa = 4.2NLDD35 pKa = 3.61PDD37 pKa = 3.56SWIEE41 pKa = 3.78TTYY44 pKa = 10.69YY45 pKa = 10.86GPISITITNDD55 pKa = 2.76VTNSQITFTNGASYY69 pKa = 10.28TPVSPTAGTTNQPIGGFSLQADD91 pKa = 4.55FPGASLTSVTISTTGTRR108 pKa = 11.84SGITNLKK115 pKa = 9.53LWSSTDD121 pKa = 3.01ATFNASSDD129 pKa = 3.63TLLNSQSDD137 pKa = 3.9NASVTFSGFSSTISTAGSYY156 pKa = 10.58YY157 pKa = 10.48FVTADD162 pKa = 4.18LSTSASGSIALTIGSVSNLTFSGGAIASGFTNAPLSSGTITIILIRR208 pKa = 11.84EE209 pKa = 3.9MNITGNGVTILNGDD223 pKa = 4.12TSPAASDD230 pKa = 3.37HH231 pKa = 6.37TDD233 pKa = 3.54FGSVDD238 pKa = 3.23IASGSVVHH246 pKa = 6.71TFTIQNNGAEE256 pKa = 4.42ALSLTGSSPFVAISGATADD275 pKa = 5.18FSLTQNPSSSIAGNSSTTFEE295 pKa = 4.03ITFNPASTGLKK306 pKa = 9.8SAVISIANDD315 pKa = 3.7DD316 pKa = 3.97GDD318 pKa = 4.18EE319 pKa = 4.1NPYY322 pKa = 9.89TFTIQGTGTAVPNMSVSGNAMQITNGDD349 pKa = 3.81IEE351 pKa = 4.5PSEE354 pKa = 5.06SDD356 pKa = 3.28DD357 pKa = 3.64TDD359 pKa = 3.78FGSVNIAGDD368 pKa = 4.27PIEE371 pKa = 4.35HH372 pKa = 6.78TFTIANNGSASLTLTGSSPYY392 pKa = 10.67VSLTGDD398 pKa = 3.56TEE400 pKa = 5.47DD401 pKa = 3.76FTLTQLPASNIAAGGGTTTFKK422 pKa = 9.92ITFKK426 pKa = 9.92PTATGNRR433 pKa = 11.84SATVSIANDD442 pKa = 3.89DD443 pKa = 3.92DD444 pKa = 4.34SKK446 pKa = 11.83NPFNFSIQGSGISVPTVTTAAASSVTKK473 pKa = 9.93TVATLGGNVTSDD485 pKa = 3.02GGSPITEE492 pKa = 4.01TGVVYY497 pKa = 10.61SSSNATPTIGGMDD510 pKa = 4.19VIQNANIDD518 pKa = 3.69LSGDD522 pKa = 3.44FSEE525 pKa = 5.52SISPLSSATHH535 pKa = 6.38YY536 pKa = 10.43YY537 pKa = 8.74FQAYY541 pKa = 6.59ATNAAGTSYY550 pKa = 11.18GGVLEE555 pKa = 4.38FTTLTSVVSILRR567 pKa = 11.84ADD569 pKa = 4.25PDD571 pKa = 3.71DD572 pKa = 3.95TNAASVRR579 pKa = 11.84WNVVFEE585 pKa = 4.42NNVTGLSASNFTLVSFGISGAAITGVSGSGTNWIVTASTGSGTGSLRR632 pKa = 11.84LGLVNDD638 pKa = 4.04NGLTPKK644 pKa = 10.56LSNPTLGGDD653 pKa = 3.91AYY655 pKa = 9.86TLDD658 pKa = 3.94LVSPSVTDD666 pKa = 3.32VDD668 pKa = 3.63TGKK671 pKa = 9.85PDD673 pKa = 3.05GTYY676 pKa = 10.99GVGSSIGIVIMFDD689 pKa = 3.09EE690 pKa = 4.4SHH692 pKa = 6.55NVDD695 pKa = 3.44GTPQLTLKK703 pKa = 8.88MDD705 pKa = 3.41SMNRR709 pKa = 11.84VIDD712 pKa = 4.74CISNEE717 pKa = 4.47HH718 pKa = 6.38IRR720 pKa = 11.84LICTYY725 pKa = 8.99TVQAGDD731 pKa = 3.56NTSDD735 pKa = 3.56LDD737 pKa = 4.12YY738 pKa = 11.62AGMTALSTNGGTIRR752 pKa = 11.84DD753 pKa = 3.79YY754 pKa = 11.22HH755 pKa = 7.8GNDD758 pKa = 4.57AILNLPWNAGTGSLAGNNAIVIDD781 pKa = 4.09TTPPEE786 pKa = 4.15ITIHH790 pKa = 6.26HH791 pKa = 7.19PGSSAAVSKK800 pKa = 9.77TITATTSDD808 pKa = 3.7GTIKK812 pKa = 10.23MSNTRR817 pKa = 11.84GTTCDD822 pKa = 2.97GSLIFTAYY830 pKa = 10.49ASQTFNAEE838 pKa = 3.55TDD840 pKa = 3.02NGIRR844 pKa = 11.84VCYY847 pKa = 9.75QAVDD851 pKa = 3.68TVNNISYY858 pKa = 10.4KK859 pKa = 10.63LSDD862 pKa = 5.22PISGIDD868 pKa = 3.3TTKK871 pKa = 9.28PTTSIIAKK879 pKa = 9.42PGNPDD884 pKa = 2.97NSASPSFTFSGEE896 pKa = 4.22DD897 pKa = 3.4PDD899 pKa = 4.5GTDD902 pKa = 3.65GSDD905 pKa = 3.15GSGIAGFEE913 pKa = 4.16CSLDD917 pKa = 3.6GEE919 pKa = 4.76PFSSCSSPKK928 pKa = 9.76TYY930 pKa = 10.92SSLGDD935 pKa = 3.6GNHH938 pKa = 5.92TFQVRR943 pKa = 11.84SVDD946 pKa = 3.45AAGNTDD952 pKa = 4.84DD953 pKa = 5.56SPEE956 pKa = 4.23SYY958 pKa = 8.6TWQIDD963 pKa = 3.37TSLPEE968 pKa = 4.05TTILSSPANPTASTDD983 pKa = 3.24AVFEE987 pKa = 4.27FEE989 pKa = 4.36GADD992 pKa = 3.43PGGSGVSGFEE1002 pKa = 4.05CSLDD1006 pKa = 3.55GEE1008 pKa = 4.6PFSDD1012 pKa = 3.81CSSPKK1017 pKa = 10.22SYY1019 pKa = 11.33SSLSDD1024 pKa = 3.39GNHH1027 pKa = 5.58TFLVRR1032 pKa = 11.84SVDD1035 pKa = 3.49AVGNADD1041 pKa = 3.55SSPAEE1046 pKa = 4.22FNWTVDD1052 pKa = 3.32TTPPEE1057 pKa = 4.07IAIHH1061 pKa = 6.17HH1062 pKa = 6.83PGSSAAVSKK1071 pKa = 9.77TITATTSDD1079 pKa = 3.7GTIKK1083 pKa = 10.23MSNTRR1088 pKa = 11.84GTTCDD1093 pKa = 3.06GSLTFTVYY1101 pKa = 10.91ASQTFNAEE1109 pKa = 3.55TDD1111 pKa = 3.02NGIRR1115 pKa = 11.84VCYY1118 pKa = 9.6QAVDD1122 pKa = 3.4AVNNISYY1129 pKa = 10.53KK1130 pKa = 10.7LSDD1133 pKa = 5.22PISGIDD1139 pKa = 3.3TTKK1142 pKa = 9.28PTTSIIAKK1150 pKa = 9.42PGNPDD1155 pKa = 2.97NSASPSFTFSGEE1167 pKa = 4.2DD1168 pKa = 3.38PDD1170 pKa = 5.4GADD1173 pKa = 3.81GSDD1176 pKa = 3.18GSGIAGFEE1184 pKa = 4.16CSLDD1188 pKa = 3.6GEE1190 pKa = 4.76PFSSCSSPKK1199 pKa = 9.76TYY1201 pKa = 10.92SSLGDD1206 pKa = 3.6GNHH1209 pKa = 5.92TFQVRR1214 pKa = 11.84SVDD1217 pKa = 3.45AAGNTDD1223 pKa = 4.84DD1224 pKa = 5.56SPEE1227 pKa = 4.23SYY1229 pKa = 8.6TWQIDD1234 pKa = 3.37TSLPEE1239 pKa = 4.05TTILSSPANLTTSTDD1254 pKa = 3.13AVFEE1258 pKa = 4.26FEE1260 pKa = 4.77GDD1262 pKa = 3.68DD1263 pKa = 3.68PGGSGIEE1270 pKa = 4.09NLEE1273 pKa = 4.11CMLDD1277 pKa = 3.26SSTFSVCTSPKK1288 pKa = 9.96EE1289 pKa = 4.09YY1290 pKa = 10.57NALPDD1295 pKa = 3.93GSHH1298 pKa = 6.17NFQVRR1303 pKa = 11.84AVDD1306 pKa = 3.32TAGNVDD1312 pKa = 4.29PSPAEE1317 pKa = 3.99INWTIDD1323 pKa = 3.03TTAPTVLISSTEE1335 pKa = 3.89SDD1337 pKa = 3.6PTNVNPIPITVQFSEE1352 pKa = 4.42PVTGFEE1358 pKa = 4.6EE1359 pKa = 4.48SDD1361 pKa = 2.95IMYY1364 pKa = 9.16HH1365 pKa = 5.71QRR1367 pKa = 11.84LPRR1370 pKa = 11.84SGQFHH1375 pKa = 6.71VFLQFDD1381 pKa = 3.88PEE1383 pKa = 4.06QRR1385 pKa = 11.84RR1386 pKa = 11.84DD1387 pKa = 3.17SYY1389 pKa = 11.32PRR1391 pKa = 11.84YY1392 pKa = 9.81SSSCRR1397 pKa = 11.84RR1398 pKa = 3.43
Molecular weight: 144.89 kDa
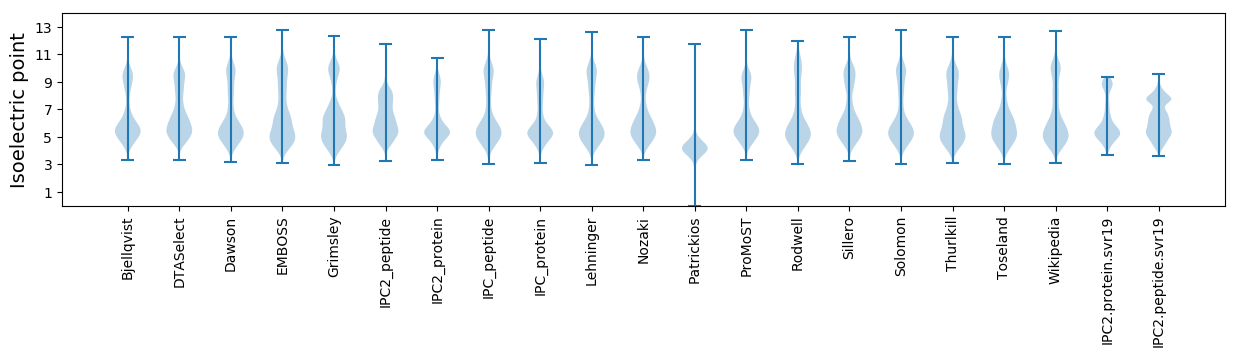
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0K8PAL5|A0A0K8PAL5_9CHLR Ribokinase OS=Flexilinea flocculi OX=1678840 GN=rbsK PE=3 SV=1
MM1 pKa = 7.55NSNTNHH7 pKa = 6.16IQTCYY12 pKa = 10.27KK13 pKa = 9.91HH14 pKa = 6.66PNRR17 pKa = 11.84QTVLRR22 pKa = 11.84CNQCGRR28 pKa = 11.84PICLDD33 pKa = 3.66CAVSTPTGYY42 pKa = 10.06RR43 pKa = 11.84CKK45 pKa = 10.67EE46 pKa = 4.23CIKK49 pKa = 9.5EE50 pKa = 3.84QQRR53 pKa = 11.84RR54 pKa = 11.84FNNSIPRR61 pKa = 11.84DD62 pKa = 3.68YY63 pKa = 11.12IFAGLIAFILGLAGSYY79 pKa = 10.15FLIVLRR85 pKa = 11.84ILSGIFALLLGPSIGMLVVKK105 pKa = 9.44LVRR108 pKa = 11.84AVTQKK113 pKa = 10.36RR114 pKa = 11.84RR115 pKa = 11.84SSTLNHH121 pKa = 5.19VTVIAAGIGAGLPLLSNVFRR141 pKa = 11.84ILQSIVVGNFRR152 pKa = 11.84LLFSNTLEE160 pKa = 4.39IGWGVAFLVLLCSVILSNMKK180 pKa = 10.04GFSIRR185 pKa = 11.84RR186 pKa = 3.66
MM1 pKa = 7.55NSNTNHH7 pKa = 6.16IQTCYY12 pKa = 10.27KK13 pKa = 9.91HH14 pKa = 6.66PNRR17 pKa = 11.84QTVLRR22 pKa = 11.84CNQCGRR28 pKa = 11.84PICLDD33 pKa = 3.66CAVSTPTGYY42 pKa = 10.06RR43 pKa = 11.84CKK45 pKa = 10.67EE46 pKa = 4.23CIKK49 pKa = 9.5EE50 pKa = 3.84QQRR53 pKa = 11.84RR54 pKa = 11.84FNNSIPRR61 pKa = 11.84DD62 pKa = 3.68YY63 pKa = 11.12IFAGLIAFILGLAGSYY79 pKa = 10.15FLIVLRR85 pKa = 11.84ILSGIFALLLGPSIGMLVVKK105 pKa = 9.44LVRR108 pKa = 11.84AVTQKK113 pKa = 10.36RR114 pKa = 11.84RR115 pKa = 11.84SSTLNHH121 pKa = 5.19VTVIAAGIGAGLPLLSNVFRR141 pKa = 11.84ILQSIVVGNFRR152 pKa = 11.84LLFSNTLEE160 pKa = 4.39IGWGVAFLVLLCSVILSNMKK180 pKa = 10.04GFSIRR185 pKa = 11.84RR186 pKa = 3.66
Molecular weight: 20.59 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
989891 |
39 |
18250 |
356.8 |
39.95 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.227 ± 0.05 | 1.219 ± 0.023 |
5.351 ± 0.047 | 6.213 ± 0.06 |
4.96 ± 0.049 | 6.811 ± 0.086 |
1.823 ± 0.024 | 8.589 ± 0.048 |
5.537 ± 0.064 | 9.699 ± 0.059 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.583 ± 0.035 | 4.614 ± 0.051 |
4.179 ± 0.043 | 3.796 ± 0.034 |
4.404 ± 0.055 | 6.787 ± 0.068 |
5.336 ± 0.091 | 6.142 ± 0.033 |
1.269 ± 0.021 | 3.461 ± 0.03 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
