
Haloarchaeobius iranensis
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Stenosarchaea group; Halobacteria; Halobacteriales; Halobacteriaceae; Haloarchaeobius
Average proteome isoelectric point is 4.88
Get precalculated fractions of proteins
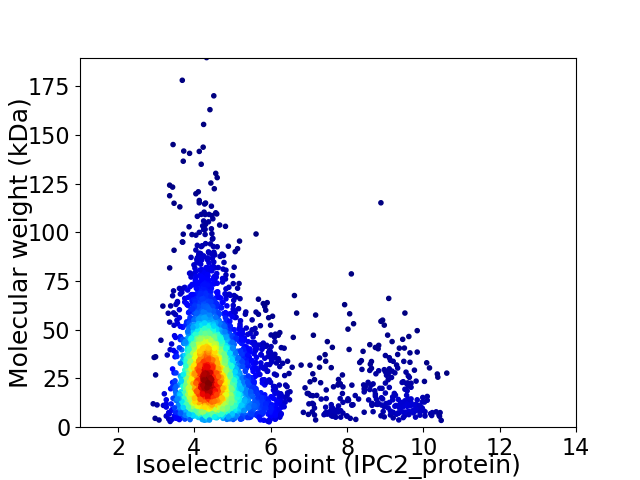
Virtual 2D-PAGE plot for 3852 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
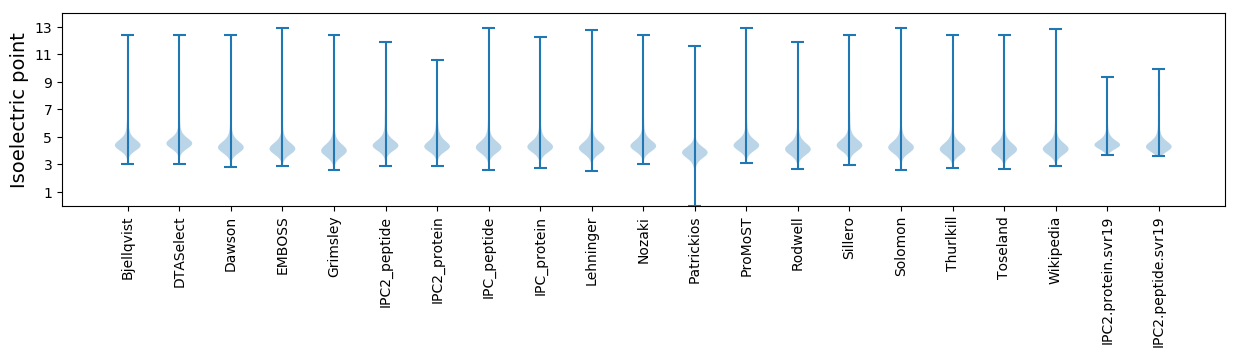
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1G9XM11|A0A1G9XM11_9EURY PD-(D/E)XK endonuclease OS=Haloarchaeobius iranensis OX=996166 GN=SAMN05192554_11130 PE=4 SV=1
MM1 pKa = 7.43VRR3 pKa = 11.84DD4 pKa = 3.76TNGVSRR10 pKa = 11.84RR11 pKa = 11.84KK12 pKa = 9.46LLKK15 pKa = 10.65GVGAGLAATTMAGEE29 pKa = 4.49VVAGEE34 pKa = 4.37GAQDD38 pKa = 3.81VIVGVSDD45 pKa = 3.73DD46 pKa = 4.84HH47 pKa = 6.84GLQTARR53 pKa = 11.84EE54 pKa = 4.08RR55 pKa = 11.84ASSVRR60 pKa = 11.84HH61 pKa = 5.6EE62 pKa = 4.72FDD64 pKa = 4.28FDD66 pKa = 5.38DD67 pKa = 4.31IGQAVAGRR75 pKa = 11.84FPAEE79 pKa = 5.21AIDD82 pKa = 4.03ALQNNPHH89 pKa = 5.03VRR91 pKa = 11.84YY92 pKa = 9.84VEE94 pKa = 3.96RR95 pKa = 11.84DD96 pKa = 3.47GEE98 pKa = 4.39YY99 pKa = 10.32HH100 pKa = 7.3ALAQTLPWGVDD111 pKa = 2.99RR112 pKa = 11.84VDD114 pKa = 4.03ADD116 pKa = 3.8VLHH119 pKa = 6.92GNGDD123 pKa = 3.83TASGADD129 pKa = 3.48IAILDD134 pKa = 3.74TGIDD138 pKa = 3.59SDD140 pKa = 5.41HH141 pKa = 7.65PDD143 pKa = 3.62LADD146 pKa = 4.16NIQGGKK152 pKa = 10.14CFTDD156 pKa = 3.5NCCGEE161 pKa = 4.25AGGGGGPFGCDD172 pKa = 2.95TNTNDD177 pKa = 5.83CPNAWDD183 pKa = 4.92DD184 pKa = 4.64DD185 pKa = 4.42NDD187 pKa = 4.45HH188 pKa = 6.37GTHH191 pKa = 6.69CAGIADD197 pKa = 3.94AVDD200 pKa = 3.41NTEE203 pKa = 3.96GVVGVSTQANLWAGKK218 pKa = 10.17VLDD221 pKa = 4.26GCGSGSLSAIAAGIEE236 pKa = 3.76WAADD240 pKa = 3.16QGFDD244 pKa = 3.67VASMSLGASSGDD256 pKa = 3.32QTLQDD261 pKa = 3.64AVQYY265 pKa = 10.41AANQGLLMVAAAGNDD280 pKa = 4.51GPCSDD285 pKa = 4.28CVGYY289 pKa = 8.77PAAYY293 pKa = 8.96PEE295 pKa = 4.5VIAVSSTASDD305 pKa = 3.9DD306 pKa = 3.88SLSDD310 pKa = 3.89FSSTGPEE317 pKa = 3.93VEE319 pKa = 4.35LAAPGTDD326 pKa = 2.67IYY328 pKa = 10.94STVPSGYY335 pKa = 8.66ATFSGTSMACPHH347 pKa = 6.06VSGAAAQVMATGATASEE364 pKa = 4.34ARR366 pKa = 11.84TILQDD371 pKa = 3.36SAEE374 pKa = 4.84DD375 pKa = 3.08IGLAGNEE382 pKa = 3.95QGYY385 pKa = 9.34GLVDD389 pKa = 3.64AEE391 pKa = 4.41NAVAAAGGGDD401 pKa = 3.91GGDD404 pKa = 3.7AAPSVSWANPAGGDD418 pKa = 3.83TVSDD422 pKa = 3.7TVSIQLSASDD432 pKa = 5.48AEE434 pKa = 4.78DD435 pKa = 3.83SDD437 pKa = 5.24DD438 pKa = 3.93SLIVEE443 pKa = 4.5WSVDD447 pKa = 3.25GGPAQTASYY456 pKa = 10.44DD457 pKa = 3.83STSGYY462 pKa = 10.84YY463 pKa = 8.76EE464 pKa = 4.55ASWDD468 pKa = 3.74STGVADD474 pKa = 5.44GDD476 pKa = 4.07HH477 pKa = 6.38TLSATATDD485 pKa = 3.72AAGNTSSSSITVTTDD500 pKa = 2.71NSSSGSTAPAIDD512 pKa = 3.78EE513 pKa = 4.17FAVTNNSNGGWARR526 pKa = 11.84FDD528 pKa = 3.64VDD530 pKa = 3.69WAVSDD535 pKa = 4.33ADD537 pKa = 3.85GDD539 pKa = 4.18LASVDD544 pKa = 4.65LVLDD548 pKa = 3.72QNGTADD554 pKa = 3.61SASDD558 pKa = 3.77SVSGSSASGSTRR570 pKa = 11.84LQDD573 pKa = 3.52KK574 pKa = 10.09KK575 pKa = 11.29GSGSYY580 pKa = 10.84DD581 pKa = 3.19VTLTVIDD588 pKa = 3.86AAGNTTSQTKK598 pKa = 8.34TVTAA602 pKa = 4.48
MM1 pKa = 7.43VRR3 pKa = 11.84DD4 pKa = 3.76TNGVSRR10 pKa = 11.84RR11 pKa = 11.84KK12 pKa = 9.46LLKK15 pKa = 10.65GVGAGLAATTMAGEE29 pKa = 4.49VVAGEE34 pKa = 4.37GAQDD38 pKa = 3.81VIVGVSDD45 pKa = 3.73DD46 pKa = 4.84HH47 pKa = 6.84GLQTARR53 pKa = 11.84EE54 pKa = 4.08RR55 pKa = 11.84ASSVRR60 pKa = 11.84HH61 pKa = 5.6EE62 pKa = 4.72FDD64 pKa = 4.28FDD66 pKa = 5.38DD67 pKa = 4.31IGQAVAGRR75 pKa = 11.84FPAEE79 pKa = 5.21AIDD82 pKa = 4.03ALQNNPHH89 pKa = 5.03VRR91 pKa = 11.84YY92 pKa = 9.84VEE94 pKa = 3.96RR95 pKa = 11.84DD96 pKa = 3.47GEE98 pKa = 4.39YY99 pKa = 10.32HH100 pKa = 7.3ALAQTLPWGVDD111 pKa = 2.99RR112 pKa = 11.84VDD114 pKa = 4.03ADD116 pKa = 3.8VLHH119 pKa = 6.92GNGDD123 pKa = 3.83TASGADD129 pKa = 3.48IAILDD134 pKa = 3.74TGIDD138 pKa = 3.59SDD140 pKa = 5.41HH141 pKa = 7.65PDD143 pKa = 3.62LADD146 pKa = 4.16NIQGGKK152 pKa = 10.14CFTDD156 pKa = 3.5NCCGEE161 pKa = 4.25AGGGGGPFGCDD172 pKa = 2.95TNTNDD177 pKa = 5.83CPNAWDD183 pKa = 4.92DD184 pKa = 4.64DD185 pKa = 4.42NDD187 pKa = 4.45HH188 pKa = 6.37GTHH191 pKa = 6.69CAGIADD197 pKa = 3.94AVDD200 pKa = 3.41NTEE203 pKa = 3.96GVVGVSTQANLWAGKK218 pKa = 10.17VLDD221 pKa = 4.26GCGSGSLSAIAAGIEE236 pKa = 3.76WAADD240 pKa = 3.16QGFDD244 pKa = 3.67VASMSLGASSGDD256 pKa = 3.32QTLQDD261 pKa = 3.64AVQYY265 pKa = 10.41AANQGLLMVAAAGNDD280 pKa = 4.51GPCSDD285 pKa = 4.28CVGYY289 pKa = 8.77PAAYY293 pKa = 8.96PEE295 pKa = 4.5VIAVSSTASDD305 pKa = 3.9DD306 pKa = 3.88SLSDD310 pKa = 3.89FSSTGPEE317 pKa = 3.93VEE319 pKa = 4.35LAAPGTDD326 pKa = 2.67IYY328 pKa = 10.94STVPSGYY335 pKa = 8.66ATFSGTSMACPHH347 pKa = 6.06VSGAAAQVMATGATASEE364 pKa = 4.34ARR366 pKa = 11.84TILQDD371 pKa = 3.36SAEE374 pKa = 4.84DD375 pKa = 3.08IGLAGNEE382 pKa = 3.95QGYY385 pKa = 9.34GLVDD389 pKa = 3.64AEE391 pKa = 4.41NAVAAAGGGDD401 pKa = 3.91GGDD404 pKa = 3.7AAPSVSWANPAGGDD418 pKa = 3.83TVSDD422 pKa = 3.7TVSIQLSASDD432 pKa = 5.48AEE434 pKa = 4.78DD435 pKa = 3.83SDD437 pKa = 5.24DD438 pKa = 3.93SLIVEE443 pKa = 4.5WSVDD447 pKa = 3.25GGPAQTASYY456 pKa = 10.44DD457 pKa = 3.83STSGYY462 pKa = 10.84YY463 pKa = 8.76EE464 pKa = 4.55ASWDD468 pKa = 3.74STGVADD474 pKa = 5.44GDD476 pKa = 4.07HH477 pKa = 6.38TLSATATDD485 pKa = 3.72AAGNTSSSSITVTTDD500 pKa = 2.71NSSSGSTAPAIDD512 pKa = 3.78EE513 pKa = 4.17FAVTNNSNGGWARR526 pKa = 11.84FDD528 pKa = 3.64VDD530 pKa = 3.69WAVSDD535 pKa = 4.33ADD537 pKa = 3.85GDD539 pKa = 4.18LASVDD544 pKa = 4.65LVLDD548 pKa = 3.72QNGTADD554 pKa = 3.61SASDD558 pKa = 3.77SVSGSSASGSTRR570 pKa = 11.84LQDD573 pKa = 3.52KK574 pKa = 10.09KK575 pKa = 11.29GSGSYY580 pKa = 10.84DD581 pKa = 3.19VTLTVIDD588 pKa = 3.86AAGNTTSQTKK598 pKa = 8.34TVTAA602 pKa = 4.48
Molecular weight: 60.27 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1G9ZRU3|A0A1G9ZRU3_9EURY Ribose 1 5-bisphosphate isomerase OS=Haloarchaeobius iranensis OX=996166 GN=SAMN05192554_12215 PE=3 SV=1
AA1 pKa = 7.46TPLPAGWLICISAASVSFRR20 pKa = 11.84AIEE23 pKa = 4.08TACLTKK29 pKa = 10.4RR30 pKa = 11.84KK31 pKa = 8.62RR32 pKa = 11.84TFCTTVLSRR41 pKa = 11.84FTSNVLTARR50 pKa = 11.84PYY52 pKa = 10.73AVRR55 pKa = 11.84GFNRR59 pKa = 11.84AVCDD63 pKa = 3.93SIRR66 pKa = 11.84NRR68 pKa = 11.84RR69 pKa = 11.84DD70 pKa = 2.24WDD72 pKa = 3.81FVIKK76 pKa = 10.45RR77 pKa = 11.84IEE79 pKa = 4.22CFLEE83 pKa = 2.92IWMIIDD89 pKa = 3.7AVFGLLAHH97 pKa = 6.91DD98 pKa = 4.72WYY100 pKa = 10.28RR101 pKa = 11.84TDD103 pKa = 3.77SRR105 pKa = 11.84EE106 pKa = 3.91ASNYY110 pKa = 7.72PQLIDD115 pKa = 3.34VRR117 pKa = 11.84KK118 pKa = 8.84IRR120 pKa = 11.84EE121 pKa = 3.88DD122 pKa = 3.38SRR124 pKa = 11.84VQDD127 pKa = 3.52ARR129 pKa = 11.84IRR131 pKa = 11.84WTITTGHH138 pKa = 6.54FADD141 pKa = 5.14DD142 pKa = 3.84LSLPEE147 pKa = 4.45SSSSNSSSLAASTSYY162 pKa = 10.87AASSSSTTPRR172 pKa = 11.84RR173 pKa = 11.84SSTPCSRR180 pKa = 11.84RR181 pKa = 11.84MSRR184 pKa = 11.84IVHH187 pKa = 6.66RR188 pKa = 11.84STSASLAASPSVIRR202 pKa = 11.84SRR204 pKa = 11.84SS205 pKa = 3.26
AA1 pKa = 7.46TPLPAGWLICISAASVSFRR20 pKa = 11.84AIEE23 pKa = 4.08TACLTKK29 pKa = 10.4RR30 pKa = 11.84KK31 pKa = 8.62RR32 pKa = 11.84TFCTTVLSRR41 pKa = 11.84FTSNVLTARR50 pKa = 11.84PYY52 pKa = 10.73AVRR55 pKa = 11.84GFNRR59 pKa = 11.84AVCDD63 pKa = 3.93SIRR66 pKa = 11.84NRR68 pKa = 11.84RR69 pKa = 11.84DD70 pKa = 2.24WDD72 pKa = 3.81FVIKK76 pKa = 10.45RR77 pKa = 11.84IEE79 pKa = 4.22CFLEE83 pKa = 2.92IWMIIDD89 pKa = 3.7AVFGLLAHH97 pKa = 6.91DD98 pKa = 4.72WYY100 pKa = 10.28RR101 pKa = 11.84TDD103 pKa = 3.77SRR105 pKa = 11.84EE106 pKa = 3.91ASNYY110 pKa = 7.72PQLIDD115 pKa = 3.34VRR117 pKa = 11.84KK118 pKa = 8.84IRR120 pKa = 11.84EE121 pKa = 3.88DD122 pKa = 3.38SRR124 pKa = 11.84VQDD127 pKa = 3.52ARR129 pKa = 11.84IRR131 pKa = 11.84WTITTGHH138 pKa = 6.54FADD141 pKa = 5.14DD142 pKa = 3.84LSLPEE147 pKa = 4.45SSSSNSSSLAASTSYY162 pKa = 10.87AASSSSTTPRR172 pKa = 11.84RR173 pKa = 11.84SSTPCSRR180 pKa = 11.84RR181 pKa = 11.84MSRR184 pKa = 11.84IVHH187 pKa = 6.66RR188 pKa = 11.84STSASLAASPSVIRR202 pKa = 11.84SRR204 pKa = 11.84SS205 pKa = 3.26
Molecular weight: 22.95 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1096490 |
29 |
1715 |
284.7 |
30.82 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.121 ± 0.055 | 0.756 ± 0.014 |
8.566 ± 0.057 | 8.302 ± 0.06 |
3.296 ± 0.025 | 8.779 ± 0.036 |
2.022 ± 0.02 | 3.363 ± 0.031 |
1.666 ± 0.024 | 8.9 ± 0.055 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.732 ± 0.019 | 2.247 ± 0.023 |
4.704 ± 0.025 | 2.542 ± 0.024 |
6.53 ± 0.041 | 5.476 ± 0.034 |
6.559 ± 0.036 | 9.602 ± 0.051 |
1.172 ± 0.016 | 2.665 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
