
Naumannella halotolerans
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Propionibacteriales; Propionibacteriaceae; Naumannella
Average proteome isoelectric point is 6.01
Get precalculated fractions of proteins
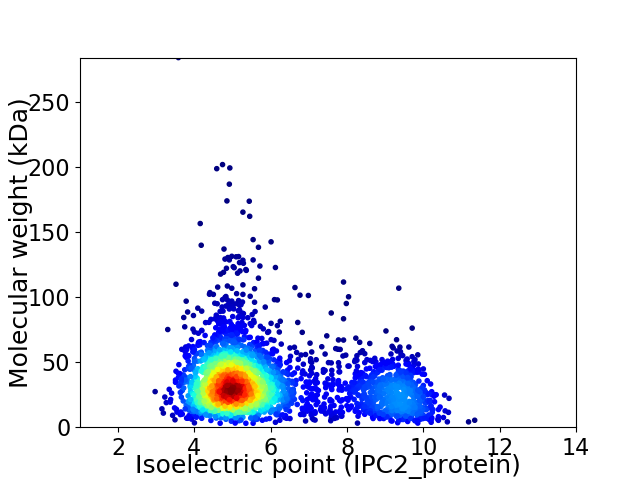
Virtual 2D-PAGE plot for 3108 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4R7J8X6|A0A4R7J8X6_9ACTN DNA-binding NarL/FixJ family response regulator OS=Naumannella halotolerans OX=993414 GN=CLV29_1600 PE=4 SV=1
MM1 pKa = 7.05VQGTSQAVPEE11 pKa = 4.5RR12 pKa = 11.84NHH14 pKa = 4.14VTNRR18 pKa = 11.84ARR20 pKa = 11.84STTRR24 pKa = 11.84IAAGLAALSLGLAACSGGGSSDD46 pKa = 3.75GEE48 pKa = 4.18GDD50 pKa = 3.29GGEE53 pKa = 4.44DD54 pKa = 3.38LTVTSSISQDD64 pKa = 2.94VDD66 pKa = 3.29TLLPMDD72 pKa = 5.17ANVGDD77 pKa = 4.67NIAVLDD83 pKa = 4.21VIYY86 pKa = 10.69DD87 pKa = 3.48GLVRR91 pKa = 11.84YY92 pKa = 9.8DD93 pKa = 3.84PEE95 pKa = 4.16TTEE98 pKa = 4.31PYY100 pKa = 10.01PYY102 pKa = 10.09LAEE105 pKa = 5.02EE106 pKa = 4.21ITTTDD111 pKa = 3.29NQVWTIKK118 pKa = 10.0IKK120 pKa = 10.74PGTTFHH126 pKa = 6.7NGEE129 pKa = 4.29PVDD132 pKa = 3.91AEE134 pKa = 3.99AFARR138 pKa = 11.84AWNYY142 pKa = 9.76AAYY145 pKa = 9.89GPNAMGNNYY154 pKa = 9.21FFEE157 pKa = 6.25RR158 pKa = 11.84IEE160 pKa = 5.34GYY162 pKa = 11.04DD163 pKa = 3.68DD164 pKa = 3.37MQGEE168 pKa = 4.48YY169 pKa = 10.83EE170 pKa = 4.16EE171 pKa = 6.46ADD173 pKa = 3.86DD174 pKa = 4.14GTVTVDD180 pKa = 3.46EE181 pKa = 5.05EE182 pKa = 4.13PAADD186 pKa = 3.88EE187 pKa = 4.26LSGLEE192 pKa = 4.09VVDD195 pKa = 4.21PQTLQVTLNAPFVGFDD211 pKa = 3.21TMLGYY216 pKa = 9.18TGFFPIAQACLDD228 pKa = 4.89DD229 pKa = 5.88IEE231 pKa = 6.12ACTTKK236 pKa = 10.61PIGNGPFQVEE246 pKa = 4.57EE247 pKa = 4.22WSQGQSLTATKK258 pKa = 10.48YY259 pKa = 10.87ADD261 pKa = 3.44YY262 pKa = 10.55TGAEE266 pKa = 4.19TPNYY270 pKa = 10.42DD271 pKa = 3.37RR272 pKa = 11.84IEE274 pKa = 4.02WVEE277 pKa = 3.79YY278 pKa = 10.69AGGSEE283 pKa = 4.1WPDD286 pKa = 3.68FQAGLMDD293 pKa = 4.78LAEE296 pKa = 4.55APSANFTQAYY306 pKa = 8.85DD307 pKa = 3.62DD308 pKa = 4.48PALQRR313 pKa = 11.84VEE315 pKa = 4.52RR316 pKa = 11.84PTAALTYY323 pKa = 10.54LSFPLYY329 pKa = 10.58DD330 pKa = 4.64DD331 pKa = 5.74GPWTDD336 pKa = 2.72IEE338 pKa = 4.27FRR340 pKa = 11.84KK341 pKa = 9.78ALSMAIDD348 pKa = 3.81RR349 pKa = 11.84QAVIDD354 pKa = 4.22AVLPGQRR361 pKa = 11.84VPADD365 pKa = 3.2SWVMPDD371 pKa = 3.71GVPGGVAGTCGEE383 pKa = 4.1ACTFNPEE390 pKa = 3.62AAKK393 pKa = 10.42AALEE397 pKa = 4.16AAGGWPEE404 pKa = 4.27GEE406 pKa = 4.32TMTIHH411 pKa = 6.89LGVDD415 pKa = 3.22EE416 pKa = 4.65TEE418 pKa = 4.11QEE420 pKa = 4.05LFQAIGDD427 pKa = 4.04QITLNLGIPYY437 pKa = 9.91EE438 pKa = 4.5LDD440 pKa = 3.21PTPDD444 pKa = 3.13FFDD447 pKa = 3.47RR448 pKa = 11.84RR449 pKa = 11.84SNRR452 pKa = 11.84DD453 pKa = 2.69FSGPFRR459 pKa = 11.84SNWFPDD465 pKa = 3.37YY466 pKa = 10.55PLNEE470 pKa = 4.4NYY472 pKa = 10.41LGPVYY477 pKa = 10.72ASDD480 pKa = 4.26DD481 pKa = 3.83AEE483 pKa = 4.1EE484 pKa = 4.87GNTNFGYY491 pKa = 10.98YY492 pKa = 10.24NEE494 pKa = 5.12DD495 pKa = 3.45FNNAIADD502 pKa = 3.86GDD504 pKa = 4.15SAATLDD510 pKa = 3.97EE511 pKa = 4.83AVTNYY516 pKa = 10.19QEE518 pKa = 4.31AEE520 pKa = 4.18RR521 pKa = 11.84ILAEE525 pKa = 4.65DD526 pKa = 5.04FPTAPLFYY534 pKa = 9.44TNVVLFHH541 pKa = 6.93SDD543 pKa = 2.79RR544 pKa = 11.84VSNVVIDD551 pKa = 4.28PFSGRR556 pKa = 11.84AKK558 pKa = 9.63MRR560 pKa = 11.84LLEE563 pKa = 4.15VTGG566 pKa = 3.81
MM1 pKa = 7.05VQGTSQAVPEE11 pKa = 4.5RR12 pKa = 11.84NHH14 pKa = 4.14VTNRR18 pKa = 11.84ARR20 pKa = 11.84STTRR24 pKa = 11.84IAAGLAALSLGLAACSGGGSSDD46 pKa = 3.75GEE48 pKa = 4.18GDD50 pKa = 3.29GGEE53 pKa = 4.44DD54 pKa = 3.38LTVTSSISQDD64 pKa = 2.94VDD66 pKa = 3.29TLLPMDD72 pKa = 5.17ANVGDD77 pKa = 4.67NIAVLDD83 pKa = 4.21VIYY86 pKa = 10.69DD87 pKa = 3.48GLVRR91 pKa = 11.84YY92 pKa = 9.8DD93 pKa = 3.84PEE95 pKa = 4.16TTEE98 pKa = 4.31PYY100 pKa = 10.01PYY102 pKa = 10.09LAEE105 pKa = 5.02EE106 pKa = 4.21ITTTDD111 pKa = 3.29NQVWTIKK118 pKa = 10.0IKK120 pKa = 10.74PGTTFHH126 pKa = 6.7NGEE129 pKa = 4.29PVDD132 pKa = 3.91AEE134 pKa = 3.99AFARR138 pKa = 11.84AWNYY142 pKa = 9.76AAYY145 pKa = 9.89GPNAMGNNYY154 pKa = 9.21FFEE157 pKa = 6.25RR158 pKa = 11.84IEE160 pKa = 5.34GYY162 pKa = 11.04DD163 pKa = 3.68DD164 pKa = 3.37MQGEE168 pKa = 4.48YY169 pKa = 10.83EE170 pKa = 4.16EE171 pKa = 6.46ADD173 pKa = 3.86DD174 pKa = 4.14GTVTVDD180 pKa = 3.46EE181 pKa = 5.05EE182 pKa = 4.13PAADD186 pKa = 3.88EE187 pKa = 4.26LSGLEE192 pKa = 4.09VVDD195 pKa = 4.21PQTLQVTLNAPFVGFDD211 pKa = 3.21TMLGYY216 pKa = 9.18TGFFPIAQACLDD228 pKa = 4.89DD229 pKa = 5.88IEE231 pKa = 6.12ACTTKK236 pKa = 10.61PIGNGPFQVEE246 pKa = 4.57EE247 pKa = 4.22WSQGQSLTATKK258 pKa = 10.48YY259 pKa = 10.87ADD261 pKa = 3.44YY262 pKa = 10.55TGAEE266 pKa = 4.19TPNYY270 pKa = 10.42DD271 pKa = 3.37RR272 pKa = 11.84IEE274 pKa = 4.02WVEE277 pKa = 3.79YY278 pKa = 10.69AGGSEE283 pKa = 4.1WPDD286 pKa = 3.68FQAGLMDD293 pKa = 4.78LAEE296 pKa = 4.55APSANFTQAYY306 pKa = 8.85DD307 pKa = 3.62DD308 pKa = 4.48PALQRR313 pKa = 11.84VEE315 pKa = 4.52RR316 pKa = 11.84PTAALTYY323 pKa = 10.54LSFPLYY329 pKa = 10.58DD330 pKa = 4.64DD331 pKa = 5.74GPWTDD336 pKa = 2.72IEE338 pKa = 4.27FRR340 pKa = 11.84KK341 pKa = 9.78ALSMAIDD348 pKa = 3.81RR349 pKa = 11.84QAVIDD354 pKa = 4.22AVLPGQRR361 pKa = 11.84VPADD365 pKa = 3.2SWVMPDD371 pKa = 3.71GVPGGVAGTCGEE383 pKa = 4.1ACTFNPEE390 pKa = 3.62AAKK393 pKa = 10.42AALEE397 pKa = 4.16AAGGWPEE404 pKa = 4.27GEE406 pKa = 4.32TMTIHH411 pKa = 6.89LGVDD415 pKa = 3.22EE416 pKa = 4.65TEE418 pKa = 4.11QEE420 pKa = 4.05LFQAIGDD427 pKa = 4.04QITLNLGIPYY437 pKa = 9.91EE438 pKa = 4.5LDD440 pKa = 3.21PTPDD444 pKa = 3.13FFDD447 pKa = 3.47RR448 pKa = 11.84RR449 pKa = 11.84SNRR452 pKa = 11.84DD453 pKa = 2.69FSGPFRR459 pKa = 11.84SNWFPDD465 pKa = 3.37YY466 pKa = 10.55PLNEE470 pKa = 4.4NYY472 pKa = 10.41LGPVYY477 pKa = 10.72ASDD480 pKa = 4.26DD481 pKa = 3.83AEE483 pKa = 4.1EE484 pKa = 4.87GNTNFGYY491 pKa = 10.98YY492 pKa = 10.24NEE494 pKa = 5.12DD495 pKa = 3.45FNNAIADD502 pKa = 3.86GDD504 pKa = 4.15SAATLDD510 pKa = 3.97EE511 pKa = 4.83AVTNYY516 pKa = 10.19QEE518 pKa = 4.31AEE520 pKa = 4.18RR521 pKa = 11.84ILAEE525 pKa = 4.65DD526 pKa = 5.04FPTAPLFYY534 pKa = 9.44TNVVLFHH541 pKa = 6.93SDD543 pKa = 2.79RR544 pKa = 11.84VSNVVIDD551 pKa = 4.28PFSGRR556 pKa = 11.84AKK558 pKa = 9.63MRR560 pKa = 11.84LLEE563 pKa = 4.15VTGG566 pKa = 3.81
Molecular weight: 61.57 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4R7J1X9|A0A4R7J1X9_9ACTN DNA-binding IclR family transcriptional regulator OS=Naumannella halotolerans OX=993414 GN=CLV29_2584 PE=4 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.01RR4 pKa = 11.84TFQPSNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84SRR15 pKa = 11.84THH17 pKa = 6.18GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84SILAARR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 10.05GRR40 pKa = 11.84TKK42 pKa = 10.74LAGG45 pKa = 3.42
MM1 pKa = 7.69SKK3 pKa = 9.01RR4 pKa = 11.84TFQPSNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84SRR15 pKa = 11.84THH17 pKa = 6.18GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84SILAARR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 10.05GRR40 pKa = 11.84TKK42 pKa = 10.74LAGG45 pKa = 3.42
Molecular weight: 5.34 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1020358 |
27 |
2777 |
328.3 |
35.25 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.528 ± 0.05 | 0.694 ± 0.013 |
6.236 ± 0.037 | 5.99 ± 0.04 |
2.885 ± 0.026 | 9.035 ± 0.041 |
1.969 ± 0.017 | 4.403 ± 0.03 |
2.016 ± 0.029 | 10.225 ± 0.054 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.892 ± 0.014 | 2.078 ± 0.021 |
5.736 ± 0.036 | 3.309 ± 0.022 |
7.295 ± 0.051 | 5.719 ± 0.034 |
6.017 ± 0.038 | 8.549 ± 0.04 |
1.52 ± 0.02 | 1.907 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
