
Microbotryum intermedium
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Basidiomycota; Pucciniomycotina; Microbotryomycetes; Microbotryales; Microbotryaceae; Microbotryum
Average proteome isoelectric point is 6.89
Get precalculated fractions of proteins
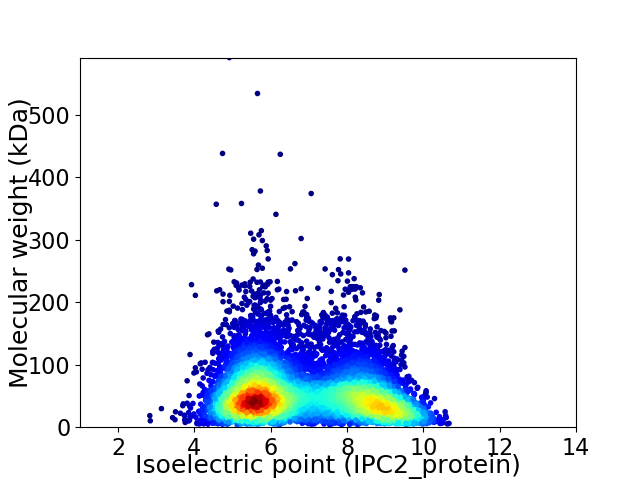
Virtual 2D-PAGE plot for 8013 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A238F381|A0A238F381_9BASI BQ2448_5118 protein OS=Microbotryum intermedium OX=269621 GN=BQ2448_5118 PE=4 SV=1
MM1 pKa = 7.65ACNTVNMFASACSSPTVVTACIGPYY26 pKa = 8.91TVSADD31 pKa = 3.48KK32 pKa = 10.71KK33 pKa = 9.45SCVLGTAWSIYY44 pKa = 9.74LLSSIPDD51 pKa = 3.38TPYY54 pKa = 10.31QSNIRR59 pKa = 11.84SPGACGNRR67 pKa = 11.84AVAKK71 pKa = 8.78GAQVAVFDD79 pKa = 4.54SGSLTCYY86 pKa = 10.75ASTANNSVSTMANLEE101 pKa = 3.95WDD103 pKa = 4.03LDD105 pKa = 3.96STTIVYY111 pKa = 7.55GTCKK115 pKa = 10.12KK116 pKa = 10.9NNVLNPAARR125 pKa = 11.84GRR127 pKa = 11.84CHH129 pKa = 7.13DD130 pKa = 4.41VILTTSACNLVADD143 pKa = 4.94KK144 pKa = 9.53STCSTLFAPCEE155 pKa = 3.92GDD157 pKa = 3.39QFRR160 pKa = 11.84NYY162 pKa = 10.95NGICQDD168 pKa = 3.4CDD170 pKa = 3.24EE171 pKa = 5.29VYY173 pKa = 10.92DD174 pKa = 4.48GALTCDD180 pKa = 3.4IFNGATSCIAGDD192 pKa = 3.71YY193 pKa = 7.52LTNGTCSSCSDD204 pKa = 4.33FDD206 pKa = 4.57PNAANCSADD215 pKa = 3.46GTVDD219 pKa = 3.11SCNDD223 pKa = 2.79GWVLSGNTCIADD235 pKa = 3.7TSSVPQIGLDD245 pKa = 4.0TAWSYY250 pKa = 11.92YY251 pKa = 9.11PDD253 pKa = 3.54SSISNLAALAVFNTNSTDD271 pKa = 3.43QYY273 pKa = 11.24DD274 pKa = 4.19CAEE277 pKa = 4.21SVYY280 pKa = 11.16DD281 pKa = 4.14SNFTMAVFGPTSQKK295 pKa = 10.71CYY297 pKa = 9.71ATNATNTLTNSIIDD311 pKa = 3.88LDD313 pKa = 4.03GASVFVIEE321 pKa = 4.68VCAANQAQVPYY332 pKa = 10.22NQGNTCYY339 pKa = 10.66DD340 pKa = 2.94ITLDD344 pKa = 4.81SEE346 pKa = 4.69SCQVNGGSCNN356 pKa = 3.45
MM1 pKa = 7.65ACNTVNMFASACSSPTVVTACIGPYY26 pKa = 8.91TVSADD31 pKa = 3.48KK32 pKa = 10.71KK33 pKa = 9.45SCVLGTAWSIYY44 pKa = 9.74LLSSIPDD51 pKa = 3.38TPYY54 pKa = 10.31QSNIRR59 pKa = 11.84SPGACGNRR67 pKa = 11.84AVAKK71 pKa = 8.78GAQVAVFDD79 pKa = 4.54SGSLTCYY86 pKa = 10.75ASTANNSVSTMANLEE101 pKa = 3.95WDD103 pKa = 4.03LDD105 pKa = 3.96STTIVYY111 pKa = 7.55GTCKK115 pKa = 10.12KK116 pKa = 10.9NNVLNPAARR125 pKa = 11.84GRR127 pKa = 11.84CHH129 pKa = 7.13DD130 pKa = 4.41VILTTSACNLVADD143 pKa = 4.94KK144 pKa = 9.53STCSTLFAPCEE155 pKa = 3.92GDD157 pKa = 3.39QFRR160 pKa = 11.84NYY162 pKa = 10.95NGICQDD168 pKa = 3.4CDD170 pKa = 3.24EE171 pKa = 5.29VYY173 pKa = 10.92DD174 pKa = 4.48GALTCDD180 pKa = 3.4IFNGATSCIAGDD192 pKa = 3.71YY193 pKa = 7.52LTNGTCSSCSDD204 pKa = 4.33FDD206 pKa = 4.57PNAANCSADD215 pKa = 3.46GTVDD219 pKa = 3.11SCNDD223 pKa = 2.79GWVLSGNTCIADD235 pKa = 3.7TSSVPQIGLDD245 pKa = 4.0TAWSYY250 pKa = 11.92YY251 pKa = 9.11PDD253 pKa = 3.54SSISNLAALAVFNTNSTDD271 pKa = 3.43QYY273 pKa = 11.24DD274 pKa = 4.19CAEE277 pKa = 4.21SVYY280 pKa = 11.16DD281 pKa = 4.14SNFTMAVFGPTSQKK295 pKa = 10.71CYY297 pKa = 9.71ATNATNTLTNSIIDD311 pKa = 3.88LDD313 pKa = 4.03GASVFVIEE321 pKa = 4.68VCAANQAQVPYY332 pKa = 10.22NQGNTCYY339 pKa = 10.66DD340 pKa = 2.94ITLDD344 pKa = 4.81SEE346 pKa = 4.69SCQVNGGSCNN356 pKa = 3.45
Molecular weight: 37.27 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A238F5F9|A0A238F5F9_9BASI BQ2448_5837 protein OS=Microbotryum intermedium OX=269621 GN=BQ2448_5837 PE=4 SV=1
MM1 pKa = 8.03RR2 pKa = 11.84SRR4 pKa = 11.84NPSSIRR10 pKa = 11.84LDD12 pKa = 4.18LIFGLYY18 pKa = 10.11LDD20 pKa = 4.54IVAGLSDD27 pKa = 3.47VRR29 pKa = 11.84IRR31 pKa = 11.84LLAANQPHH39 pKa = 7.61PFTLRR44 pKa = 11.84KK45 pKa = 9.42PPPNQHH51 pKa = 5.45RR52 pKa = 11.84RR53 pKa = 11.84PWIWIWWAYY62 pKa = 9.23FCHH65 pKa = 6.84PPPKK69 pKa = 9.07WYY71 pKa = 9.65HH72 pKa = 5.91AANHH76 pKa = 5.83VLFFPAGHH84 pKa = 7.27GIQLSVNPTLFRR96 pKa = 11.84GPDD99 pKa = 3.8GEE101 pKa = 4.81VMTPTSFIDD110 pKa = 3.48ASKK113 pKa = 10.32RR114 pKa = 11.84RR115 pKa = 11.84HH116 pKa = 5.8AFVFPPIIPEE126 pKa = 4.0GHH128 pKa = 5.45QKK130 pKa = 9.92IFFLTEE136 pKa = 3.75QRR138 pKa = 11.84WKK140 pKa = 10.58SWWKK144 pKa = 9.48ALRR147 pKa = 11.84KK148 pKa = 8.97IRR150 pKa = 11.84RR151 pKa = 11.84VHH153 pKa = 7.13SDD155 pKa = 3.32AEE157 pKa = 4.4DD158 pKa = 3.65TAHH161 pKa = 7.14LLSLGSLHH169 pKa = 7.08PGVQLSSATHH179 pKa = 5.28SQLNNRR185 pKa = 11.84SASYY189 pKa = 10.56VMCLSEE195 pKa = 4.87APEE198 pKa = 4.37SLPHH202 pKa = 6.36LAVGCTFARR211 pKa = 11.84RR212 pKa = 11.84LWAALSPSPHH222 pKa = 7.21PIFADD227 pKa = 4.87FVCPLVSRR235 pKa = 11.84SEE237 pKa = 3.86RR238 pKa = 11.84RR239 pKa = 11.84IVEE242 pKa = 3.64LRR244 pKa = 11.84ILFFHH249 pKa = 6.86SIWKK253 pKa = 9.43LSRR256 pKa = 11.84RR257 pKa = 11.84RR258 pKa = 11.84RR259 pKa = 11.84FSSQPLEE266 pKa = 4.82PITEE270 pKa = 4.29AEE272 pKa = 4.13LEE274 pKa = 4.14EE275 pKa = 4.41LRR277 pKa = 11.84GSIQGCTGLYY287 pKa = 9.97NISKK291 pKa = 10.46GGFSGYY297 pKa = 9.91GRR299 pKa = 11.84LWLKK303 pKa = 10.73GRR305 pKa = 4.01
MM1 pKa = 8.03RR2 pKa = 11.84SRR4 pKa = 11.84NPSSIRR10 pKa = 11.84LDD12 pKa = 4.18LIFGLYY18 pKa = 10.11LDD20 pKa = 4.54IVAGLSDD27 pKa = 3.47VRR29 pKa = 11.84IRR31 pKa = 11.84LLAANQPHH39 pKa = 7.61PFTLRR44 pKa = 11.84KK45 pKa = 9.42PPPNQHH51 pKa = 5.45RR52 pKa = 11.84RR53 pKa = 11.84PWIWIWWAYY62 pKa = 9.23FCHH65 pKa = 6.84PPPKK69 pKa = 9.07WYY71 pKa = 9.65HH72 pKa = 5.91AANHH76 pKa = 5.83VLFFPAGHH84 pKa = 7.27GIQLSVNPTLFRR96 pKa = 11.84GPDD99 pKa = 3.8GEE101 pKa = 4.81VMTPTSFIDD110 pKa = 3.48ASKK113 pKa = 10.32RR114 pKa = 11.84RR115 pKa = 11.84HH116 pKa = 5.8AFVFPPIIPEE126 pKa = 4.0GHH128 pKa = 5.45QKK130 pKa = 9.92IFFLTEE136 pKa = 3.75QRR138 pKa = 11.84WKK140 pKa = 10.58SWWKK144 pKa = 9.48ALRR147 pKa = 11.84KK148 pKa = 8.97IRR150 pKa = 11.84RR151 pKa = 11.84VHH153 pKa = 7.13SDD155 pKa = 3.32AEE157 pKa = 4.4DD158 pKa = 3.65TAHH161 pKa = 7.14LLSLGSLHH169 pKa = 7.08PGVQLSSATHH179 pKa = 5.28SQLNNRR185 pKa = 11.84SASYY189 pKa = 10.56VMCLSEE195 pKa = 4.87APEE198 pKa = 4.37SLPHH202 pKa = 6.36LAVGCTFARR211 pKa = 11.84RR212 pKa = 11.84LWAALSPSPHH222 pKa = 7.21PIFADD227 pKa = 4.87FVCPLVSRR235 pKa = 11.84SEE237 pKa = 3.86RR238 pKa = 11.84RR239 pKa = 11.84IVEE242 pKa = 3.64LRR244 pKa = 11.84ILFFHH249 pKa = 6.86SIWKK253 pKa = 9.43LSRR256 pKa = 11.84RR257 pKa = 11.84RR258 pKa = 11.84RR259 pKa = 11.84FSSQPLEE266 pKa = 4.82PITEE270 pKa = 4.29AEE272 pKa = 4.13LEE274 pKa = 4.14EE275 pKa = 4.41LRR277 pKa = 11.84GSIQGCTGLYY287 pKa = 9.97NISKK291 pKa = 10.46GGFSGYY297 pKa = 9.91GRR299 pKa = 11.84LWLKK303 pKa = 10.73GRR305 pKa = 4.01
Molecular weight: 35.0 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4386583 |
34 |
5376 |
547.4 |
59.93 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.405 ± 0.026 | 1.162 ± 0.014 |
5.521 ± 0.019 | 5.811 ± 0.023 |
3.447 ± 0.015 | 6.762 ± 0.023 |
2.523 ± 0.012 | 4.275 ± 0.015 |
4.493 ± 0.025 | 9.21 ± 0.029 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.919 ± 0.009 | 3.124 ± 0.012 |
6.459 ± 0.032 | 3.757 ± 0.018 |
6.493 ± 0.021 | 9.56 ± 0.042 |
6.364 ± 0.019 | 6.136 ± 0.018 |
1.296 ± 0.009 | 2.282 ± 0.011 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
