
Hyunsoonleella pacifica
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Hyunsoonleella
Average proteome isoelectric point is 6.66
Get precalculated fractions of proteins
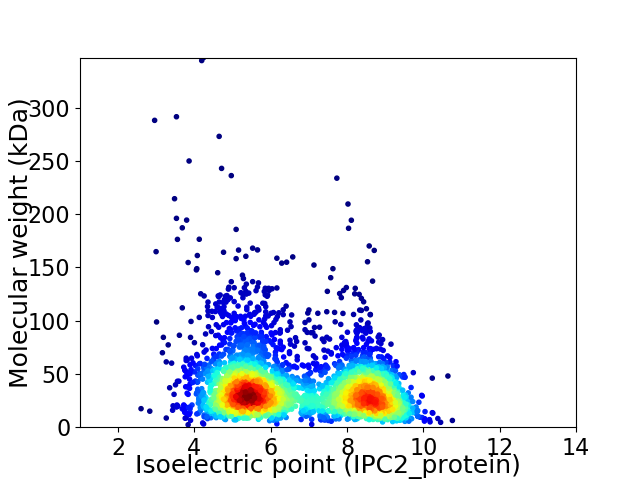
Virtual 2D-PAGE plot for 3449 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4Q9FMP5|A0A4Q9FMP5_9FLAO Alpha-L-rhamnosidase OS=Hyunsoonleella pacifica OX=1080224 GN=EYD46_16395 PE=4 SV=1
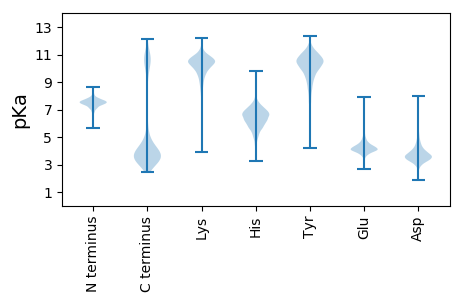
MM1 pKa = 7.34IFKK4 pKa = 10.22IIKK7 pKa = 9.12KK8 pKa = 9.53YY9 pKa = 10.18NIGVILLLSMFLALISCYY27 pKa = 9.54DD28 pKa = 3.15TGYY31 pKa = 11.23EE32 pKa = 4.06EE33 pKa = 5.3FVPPTGNVNNIQPNTLFTTTTSADD57 pKa = 3.42DD58 pKa = 3.45NLRR61 pKa = 11.84IVFRR65 pKa = 11.84SYY67 pKa = 10.09STDD70 pKa = 2.93AVSYY74 pKa = 10.43LWDD77 pKa = 4.47FGDD80 pKa = 4.46GNTSTEE86 pKa = 3.87ANPNYY91 pKa = 10.06TYY93 pKa = 10.64ATGGLYY99 pKa = 10.37AVTLNTVSSDD109 pKa = 3.19GLEE112 pKa = 4.35ANASANVSPIFVDD125 pKa = 4.81FNFSAIDD132 pKa = 3.61SEE134 pKa = 5.02VTFEE138 pKa = 4.12NLTTGAEE145 pKa = 4.1SLVWDD150 pKa = 5.02FGDD153 pKa = 4.06GEE155 pKa = 4.53SVEE158 pKa = 4.19WSIEE162 pKa = 3.99DD163 pKa = 4.31SEE165 pKa = 4.87ADD167 pKa = 3.35PDD169 pKa = 4.25FSPVYY174 pKa = 10.08SYY176 pKa = 9.63KK177 pKa = 10.07TAEE180 pKa = 4.24TFQATLTATNYY191 pKa = 10.69LGVQVSITKK200 pKa = 10.4NIEE203 pKa = 3.75GLVLSTVPDD212 pKa = 3.71FTFDD216 pKa = 3.41VSSLEE221 pKa = 4.01VQFTDD226 pKa = 3.34TSILAVSHH234 pKa = 6.25SWDD237 pKa = 3.61FGDD240 pKa = 5.51GNTSTDD246 pKa = 3.1ASPLHH251 pKa = 6.47TYY253 pKa = 7.98ATAGTYY259 pKa = 10.53DD260 pKa = 3.5VTLTTTNAAGVSRR273 pKa = 11.84SITKK277 pKa = 8.83PVPVGGIEE285 pKa = 3.9PTFKK289 pKa = 11.02VIVQNSDD296 pKa = 3.97CNVHH300 pKa = 6.1TSNTGDD306 pKa = 4.3NADD309 pKa = 3.98AWDD312 pKa = 3.94MTPNSTVVDD321 pKa = 4.47DD322 pKa = 4.44NLGTIDD328 pKa = 3.64SPYY331 pKa = 9.76RR332 pKa = 11.84ALWRR336 pKa = 11.84NDD338 pKa = 3.21EE339 pKa = 4.19LNAYY343 pKa = 9.16IDD345 pKa = 3.64ATFCTNEE352 pKa = 4.16QPGSTSDD359 pKa = 3.53GNKK362 pKa = 9.87FGPDD366 pKa = 2.9AGGGRR371 pKa = 11.84GVKK374 pKa = 10.41LSNNCRR380 pKa = 11.84RR381 pKa = 11.84LYY383 pKa = 10.48QLVAVEE389 pKa = 4.63PGVEE393 pKa = 4.03YY394 pKa = 10.23TFTIDD399 pKa = 3.3TRR401 pKa = 11.84SEE403 pKa = 3.63AAGINTEE410 pKa = 4.26VFILNNEE417 pKa = 3.87ITTEE421 pKa = 3.96EE422 pKa = 4.72AINTPAALAINTDD435 pKa = 3.78GYY437 pKa = 12.02ALIDD441 pKa = 3.46NDD443 pKa = 3.99FNSSKK448 pKa = 10.79SSSTNDD454 pKa = 2.94TFTTTTLTFEE464 pKa = 4.3ASANIAVIYY473 pKa = 10.32VRR475 pKa = 11.84ALNAIDD481 pKa = 3.79SSNEE485 pKa = 3.66VFIDD489 pKa = 4.06NIDD492 pKa = 3.83IITPGFDD499 pKa = 2.85
MM1 pKa = 7.34IFKK4 pKa = 10.22IIKK7 pKa = 9.12KK8 pKa = 9.53YY9 pKa = 10.18NIGVILLLSMFLALISCYY27 pKa = 9.54DD28 pKa = 3.15TGYY31 pKa = 11.23EE32 pKa = 4.06EE33 pKa = 5.3FVPPTGNVNNIQPNTLFTTTTSADD57 pKa = 3.42DD58 pKa = 3.45NLRR61 pKa = 11.84IVFRR65 pKa = 11.84SYY67 pKa = 10.09STDD70 pKa = 2.93AVSYY74 pKa = 10.43LWDD77 pKa = 4.47FGDD80 pKa = 4.46GNTSTEE86 pKa = 3.87ANPNYY91 pKa = 10.06TYY93 pKa = 10.64ATGGLYY99 pKa = 10.37AVTLNTVSSDD109 pKa = 3.19GLEE112 pKa = 4.35ANASANVSPIFVDD125 pKa = 4.81FNFSAIDD132 pKa = 3.61SEE134 pKa = 5.02VTFEE138 pKa = 4.12NLTTGAEE145 pKa = 4.1SLVWDD150 pKa = 5.02FGDD153 pKa = 4.06GEE155 pKa = 4.53SVEE158 pKa = 4.19WSIEE162 pKa = 3.99DD163 pKa = 4.31SEE165 pKa = 4.87ADD167 pKa = 3.35PDD169 pKa = 4.25FSPVYY174 pKa = 10.08SYY176 pKa = 9.63KK177 pKa = 10.07TAEE180 pKa = 4.24TFQATLTATNYY191 pKa = 10.69LGVQVSITKK200 pKa = 10.4NIEE203 pKa = 3.75GLVLSTVPDD212 pKa = 3.71FTFDD216 pKa = 3.41VSSLEE221 pKa = 4.01VQFTDD226 pKa = 3.34TSILAVSHH234 pKa = 6.25SWDD237 pKa = 3.61FGDD240 pKa = 5.51GNTSTDD246 pKa = 3.1ASPLHH251 pKa = 6.47TYY253 pKa = 7.98ATAGTYY259 pKa = 10.53DD260 pKa = 3.5VTLTTTNAAGVSRR273 pKa = 11.84SITKK277 pKa = 8.83PVPVGGIEE285 pKa = 3.9PTFKK289 pKa = 11.02VIVQNSDD296 pKa = 3.97CNVHH300 pKa = 6.1TSNTGDD306 pKa = 4.3NADD309 pKa = 3.98AWDD312 pKa = 3.94MTPNSTVVDD321 pKa = 4.47DD322 pKa = 4.44NLGTIDD328 pKa = 3.64SPYY331 pKa = 9.76RR332 pKa = 11.84ALWRR336 pKa = 11.84NDD338 pKa = 3.21EE339 pKa = 4.19LNAYY343 pKa = 9.16IDD345 pKa = 3.64ATFCTNEE352 pKa = 4.16QPGSTSDD359 pKa = 3.53GNKK362 pKa = 9.87FGPDD366 pKa = 2.9AGGGRR371 pKa = 11.84GVKK374 pKa = 10.41LSNNCRR380 pKa = 11.84RR381 pKa = 11.84LYY383 pKa = 10.48QLVAVEE389 pKa = 4.63PGVEE393 pKa = 4.03YY394 pKa = 10.23TFTIDD399 pKa = 3.3TRR401 pKa = 11.84SEE403 pKa = 3.63AAGINTEE410 pKa = 4.26VFILNNEE417 pKa = 3.87ITTEE421 pKa = 3.96EE422 pKa = 4.72AINTPAALAINTDD435 pKa = 3.78GYY437 pKa = 12.02ALIDD441 pKa = 3.46NDD443 pKa = 3.99FNSSKK448 pKa = 10.79SSSTNDD454 pKa = 2.94TFTTTTLTFEE464 pKa = 4.3ASANIAVIYY473 pKa = 10.32VRR475 pKa = 11.84ALNAIDD481 pKa = 3.79SSNEE485 pKa = 3.66VFIDD489 pKa = 4.06NIDD492 pKa = 3.83IITPGFDD499 pKa = 2.85
Molecular weight: 53.85 kDa
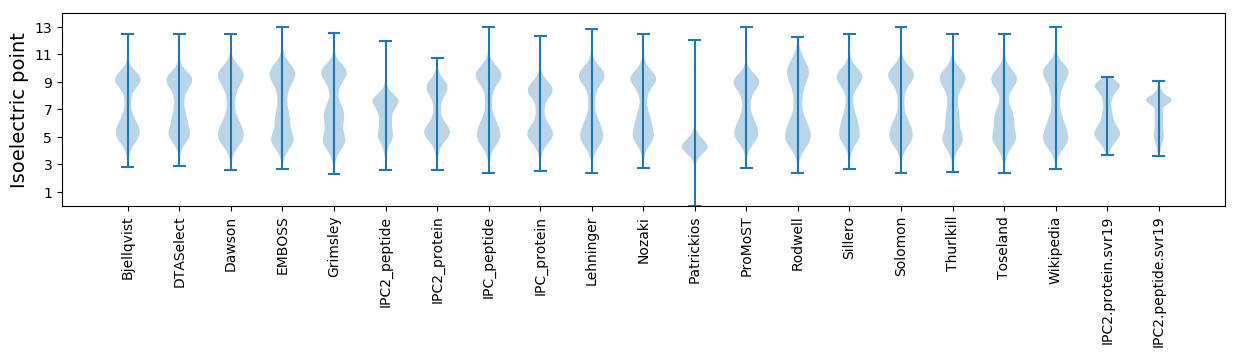
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4V2JB50|A0A4V2JB50_9FLAO 3-phosphoglycerate dehydrogenase OS=Hyunsoonleella pacifica OX=1080224 GN=EYD46_03580 PE=3 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.01RR4 pKa = 11.84TFQPSKK10 pKa = 9.13RR11 pKa = 11.84KK12 pKa = 9.48RR13 pKa = 11.84RR14 pKa = 11.84NKK16 pKa = 9.49HH17 pKa = 3.94GFRR20 pKa = 11.84EE21 pKa = 4.27RR22 pKa = 11.84MASANGRR29 pKa = 11.84KK30 pKa = 9.04VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.09GRR40 pKa = 11.84KK41 pKa = 7.97KK42 pKa = 10.62LSVSSEE48 pKa = 4.03TRR50 pKa = 11.84HH51 pKa = 5.96KK52 pKa = 10.68KK53 pKa = 9.8
MM1 pKa = 7.69SKK3 pKa = 9.01RR4 pKa = 11.84TFQPSKK10 pKa = 9.13RR11 pKa = 11.84KK12 pKa = 9.48RR13 pKa = 11.84RR14 pKa = 11.84NKK16 pKa = 9.49HH17 pKa = 3.94GFRR20 pKa = 11.84EE21 pKa = 4.27RR22 pKa = 11.84MASANGRR29 pKa = 11.84KK30 pKa = 9.04VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.09GRR40 pKa = 11.84KK41 pKa = 7.97KK42 pKa = 10.62LSVSSEE48 pKa = 4.03TRR50 pKa = 11.84HH51 pKa = 5.96KK52 pKa = 10.68KK53 pKa = 9.8
Molecular weight: 6.32 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1242761 |
22 |
3122 |
360.3 |
40.71 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.045 ± 0.041 | 0.778 ± 0.013 |
5.816 ± 0.036 | 6.38 ± 0.036 |
5.241 ± 0.033 | 6.281 ± 0.042 |
1.769 ± 0.019 | 8.105 ± 0.041 |
7.953 ± 0.068 | 9.161 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.978 ± 0.023 | 6.645 ± 0.049 |
3.292 ± 0.023 | 3.215 ± 0.022 |
3.342 ± 0.027 | 6.616 ± 0.035 |
5.985 ± 0.055 | 6.149 ± 0.028 |
1.081 ± 0.015 | 4.171 ± 0.031 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
