
Wenzhouxiangella sp. W260
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Chromatiales; Wenzhouxiangellaceae; Wenzhouxiangella; unclassified Wenzhouxiangella
Average proteome isoelectric point is 5.85
Get precalculated fractions of proteins
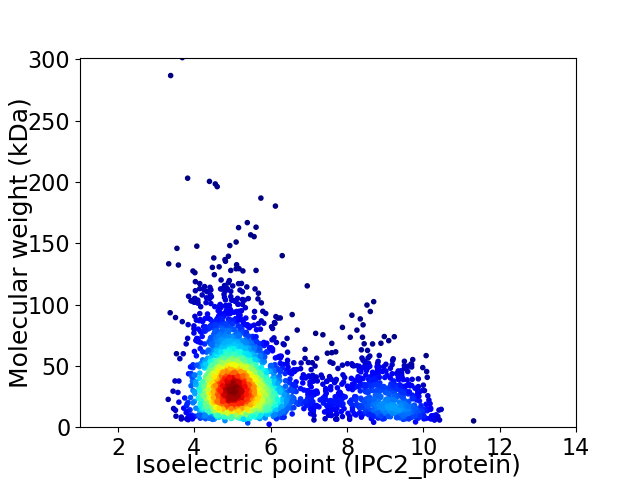
Virtual 2D-PAGE plot for 3042 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5N0TLE1|A0A5N0TLE1_9GAMM TolC family protein OS=Wenzhouxiangella sp. W260 OX=2613842 GN=F3N42_00925 PE=3 SV=1
MM1 pKa = 7.33TPTVVATASRR11 pKa = 11.84HH12 pKa = 4.91RR13 pKa = 11.84FSLALTSTLLLLAGPIEE30 pKa = 4.13AQVFINEE37 pKa = 3.83LHH39 pKa = 6.25YY40 pKa = 11.5DD41 pKa = 3.76NVSGDD46 pKa = 3.49SGEE49 pKa = 4.25MVEE52 pKa = 4.56LAGPAGTSLAGWTVALYY69 pKa = 10.57NGSASQLNVYY79 pKa = 8.45DD80 pKa = 4.58TINLSGTFADD90 pKa = 3.89QAGGMGTLAFNRR102 pKa = 11.84AGIQNGSPDD111 pKa = 3.6GLALVDD117 pKa = 3.91GSNNVVQFLSYY128 pKa = 10.79EE129 pKa = 4.41GVFTAASGPAAGMTSTDD146 pKa = 2.87IGVEE150 pKa = 4.16EE151 pKa = 5.02GSGTPVGHH159 pKa = 6.54SLQLVGAGLAYY170 pKa = 10.31DD171 pKa = 3.91DD172 pKa = 4.41FTWAEE177 pKa = 3.93AMAATPGQPNTGQDD191 pKa = 3.46FGGEE195 pKa = 3.92PSVPEE200 pKa = 4.2LLLSEE205 pKa = 5.16LVVTPTGGEE214 pKa = 3.99FIEE217 pKa = 4.19IHH219 pKa = 6.11NPGGSAVDD227 pKa = 3.98LSDD230 pKa = 5.18VYY232 pKa = 10.92LTDD235 pKa = 3.23ATFAGGNTYY244 pKa = 10.6YY245 pKa = 11.29YY246 pKa = 10.81NIVLGANAGGGGFSDD261 pKa = 3.95FHH263 pKa = 8.69ARR265 pKa = 11.84FPAGASIPAGGFQTVALAGSEE286 pKa = 4.31AFFAEE291 pKa = 4.72YY292 pKa = 10.2GVQPTYY298 pKa = 11.34EE299 pKa = 4.1LFEE302 pKa = 6.89DD303 pKa = 5.1GAADD307 pKa = 5.53AIPDD311 pKa = 3.66MRR313 pKa = 11.84EE314 pKa = 3.86ALPGSINAQGGLTNSGEE331 pKa = 4.53VVVLYY336 pKa = 10.36HH337 pKa = 6.7WDD339 pKa = 3.54EE340 pKa = 4.57SSDD343 pKa = 3.68LVADD347 pKa = 4.38LDD349 pKa = 4.13YY350 pKa = 11.4AVWGDD355 pKa = 3.27KK356 pKa = 11.17DD357 pKa = 3.7EE358 pKa = 5.72AVDD361 pKa = 3.68KK362 pKa = 11.11TGVALDD368 pKa = 4.58GPDD371 pKa = 3.57GDD373 pKa = 4.54TLVSEE378 pKa = 4.84YY379 pKa = 11.19AADD382 pKa = 3.94TPIASQDD389 pKa = 3.82VISGGAHH396 pKa = 6.41AFGEE400 pKa = 4.69SFTRR404 pKa = 11.84IDD406 pKa = 3.53FTEE409 pKa = 4.64GSEE412 pKa = 4.27TQSGGNGVNGADD424 pKa = 3.37EE425 pKa = 4.28TSEE428 pKa = 4.01NLSVTWASRR437 pKa = 11.84TPSPNSDD444 pKa = 3.88YY445 pKa = 11.31VPPTPDD451 pKa = 3.21LVITEE456 pKa = 4.12VMQNPAAVGDD466 pKa = 4.29SSGEE470 pKa = 3.59WFEE473 pKa = 3.7IHH475 pKa = 6.59NAGDD479 pKa = 3.31VDD481 pKa = 3.82VDD483 pKa = 3.68INGWTIADD491 pKa = 4.37LGSDD495 pKa = 3.4SHH497 pKa = 7.53VINAGGPLVVPAGGYY512 pKa = 8.05MVLGNNADD520 pKa = 3.69SGSNGGLVVDD530 pKa = 4.13YY531 pKa = 10.32AYY533 pKa = 11.23SGVFLANGDD542 pKa = 4.08DD543 pKa = 3.77EE544 pKa = 5.98LVLFDD549 pKa = 3.93AAGFEE554 pKa = 4.48VDD556 pKa = 3.55GVAWDD561 pKa = 4.85GGPVWPDD568 pKa = 3.01PTGASMALIDD578 pKa = 5.65LALDD582 pKa = 3.9NNDD585 pKa = 3.09GNNWCEE591 pKa = 4.0SQNPYY596 pKa = 11.1GDD598 pKa = 3.89GDD600 pKa = 3.74RR601 pKa = 11.84GTPGVANSCEE611 pKa = 3.9VLIPEE616 pKa = 5.44FGACGDD622 pKa = 3.84AATKK626 pKa = 9.92IHH628 pKa = 6.56AVQGNGLASPIAGSAGVIIEE648 pKa = 4.22GVVVGDD654 pKa = 3.5FQGGDD659 pKa = 3.14RR660 pKa = 11.84LNGFFVQEE668 pKa = 3.95EE669 pKa = 4.37SADD672 pKa = 3.74VDD674 pKa = 3.66GDD676 pKa = 3.86PATSEE681 pKa = 4.55GIFVYY686 pKa = 10.6DD687 pKa = 3.44EE688 pKa = 4.19NFGVDD693 pKa = 3.65VNVGDD698 pKa = 3.91VVRR701 pKa = 11.84VQGTVIEE708 pKa = 4.88FFDD711 pKa = 3.86LTEE714 pKa = 4.15ISPVINLVTCGTGTADD730 pKa = 3.53TQDD733 pKa = 3.14ILLPLAGPDD742 pKa = 3.24ALEE745 pKa = 4.24ALEE748 pKa = 4.35GMSVHH753 pKa = 7.03LPQTLFVTDD762 pKa = 3.68NFTLGRR768 pKa = 11.84YY769 pKa = 9.66GEE771 pKa = 4.25VGLAVGGPLDD781 pKa = 3.48IPTNVVAPGAAAQALQAQNDD801 pKa = 3.82LSRR804 pKa = 11.84IQLDD808 pKa = 4.14DD809 pKa = 3.85GSGLQNPVPVPPYY822 pKa = 9.31FAPDD826 pKa = 3.11GTLRR830 pKa = 11.84VGDD833 pKa = 3.73ATSAVTGVLGFGFGEE848 pKa = 4.4YY849 pKa = 9.7EE850 pKa = 3.74IHH852 pKa = 6.35PTDD855 pKa = 3.09VVAFSRR861 pKa = 11.84EE862 pKa = 3.79NEE864 pKa = 4.27RR865 pKa = 11.84PDD867 pKa = 4.1DD868 pKa = 4.45APDD871 pKa = 3.26VGNPLVTVAGFNVLNYY887 pKa = 7.55FTTLDD892 pKa = 4.43DD893 pKa = 3.81NTPVCGPSASMDD905 pKa = 3.22CRR907 pKa = 11.84GAEE910 pKa = 3.69NAFEE914 pKa = 4.44FEE916 pKa = 4.29RR917 pKa = 11.84QRR919 pKa = 11.84SKK921 pKa = 10.7IVAALSLLGADD932 pKa = 3.16VVGLVEE938 pKa = 4.27VEE940 pKa = 4.08NAAGDD945 pKa = 4.27GPVADD950 pKa = 5.03LVDD953 pKa = 5.33GINDD957 pKa = 3.22ILGADD962 pKa = 4.18TYY964 pKa = 11.71AYY966 pKa = 9.54IATGAIGTDD975 pKa = 4.44AIRR978 pKa = 11.84QALIYY983 pKa = 10.41KK984 pKa = 8.98PDD986 pKa = 3.28AVTPVGGFEE995 pKa = 4.46TLDD998 pKa = 3.77SSDD1001 pKa = 4.37DD1002 pKa = 3.57PAFIDD1007 pKa = 3.7TANRR1011 pKa = 11.84PALAQSFTDD1020 pKa = 4.19GSTGEE1025 pKa = 4.33VFTVAVNHH1033 pKa = 6.36LKK1035 pKa = 10.85SKK1037 pKa = 10.9GSDD1040 pKa = 3.4CDD1042 pKa = 3.77GLGDD1046 pKa = 4.84PDD1048 pKa = 4.01TGDD1051 pKa = 3.44GQGNCNLIRR1060 pKa = 11.84AAAASALATWLAADD1074 pKa = 3.8PTGSGSEE1081 pKa = 3.72RR1082 pKa = 11.84SLIIGDD1088 pKa = 4.15LNAYY1092 pKa = 9.83AKK1094 pKa = 9.95EE1095 pKa = 4.02DD1096 pKa = 4.57PIVVLKK1102 pKa = 10.93DD1103 pKa = 3.09AGFVDD1108 pKa = 5.13LVEE1111 pKa = 4.57VYY1113 pKa = 10.85NGVGYY1118 pKa = 11.02ADD1120 pKa = 3.66GAYY1123 pKa = 10.06SYY1125 pKa = 11.65GFGGQMGYY1133 pKa = 10.54LDD1135 pKa = 4.38HH1136 pKa = 7.3ALASAAFAADD1146 pKa = 3.5ASGATAWHH1154 pKa = 6.63INADD1158 pKa = 3.57EE1159 pKa = 4.33PAALDD1164 pKa = 4.14YY1165 pKa = 11.32NDD1167 pKa = 4.96HH1168 pKa = 5.99NQDD1171 pKa = 3.38VLFSPDD1177 pKa = 3.28PYY1179 pKa = 10.28RR1180 pKa = 11.84ASDD1183 pKa = 3.35HH1184 pKa = 6.7DD1185 pKa = 4.14AVVVGLFNDD1194 pKa = 3.71EE1195 pKa = 4.91DD1196 pKa = 5.36DD1197 pKa = 4.86DD1198 pKa = 5.1GVWDD1202 pKa = 5.21TIDD1205 pKa = 3.64ACPATVIPEE1214 pKa = 4.21TPPTRR1219 pKa = 11.84GLGNNRR1225 pKa = 11.84FALLDD1230 pKa = 4.1DD1231 pKa = 5.65DD1232 pKa = 5.75GVFDD1236 pKa = 3.88TTEE1239 pKa = 4.05GGGPGVSFTIHH1250 pKa = 6.38DD1251 pKa = 3.85TAGCSCEE1258 pKa = 4.12QIIAAADD1265 pKa = 3.88LGAGHH1270 pKa = 7.37AKK1272 pKa = 9.99FGCSIGEE1279 pKa = 3.74IRR1281 pKa = 11.84DD1282 pKa = 3.58WVSAQQ1287 pKa = 3.07
MM1 pKa = 7.33TPTVVATASRR11 pKa = 11.84HH12 pKa = 4.91RR13 pKa = 11.84FSLALTSTLLLLAGPIEE30 pKa = 4.13AQVFINEE37 pKa = 3.83LHH39 pKa = 6.25YY40 pKa = 11.5DD41 pKa = 3.76NVSGDD46 pKa = 3.49SGEE49 pKa = 4.25MVEE52 pKa = 4.56LAGPAGTSLAGWTVALYY69 pKa = 10.57NGSASQLNVYY79 pKa = 8.45DD80 pKa = 4.58TINLSGTFADD90 pKa = 3.89QAGGMGTLAFNRR102 pKa = 11.84AGIQNGSPDD111 pKa = 3.6GLALVDD117 pKa = 3.91GSNNVVQFLSYY128 pKa = 10.79EE129 pKa = 4.41GVFTAASGPAAGMTSTDD146 pKa = 2.87IGVEE150 pKa = 4.16EE151 pKa = 5.02GSGTPVGHH159 pKa = 6.54SLQLVGAGLAYY170 pKa = 10.31DD171 pKa = 3.91DD172 pKa = 4.41FTWAEE177 pKa = 3.93AMAATPGQPNTGQDD191 pKa = 3.46FGGEE195 pKa = 3.92PSVPEE200 pKa = 4.2LLLSEE205 pKa = 5.16LVVTPTGGEE214 pKa = 3.99FIEE217 pKa = 4.19IHH219 pKa = 6.11NPGGSAVDD227 pKa = 3.98LSDD230 pKa = 5.18VYY232 pKa = 10.92LTDD235 pKa = 3.23ATFAGGNTYY244 pKa = 10.6YY245 pKa = 11.29YY246 pKa = 10.81NIVLGANAGGGGFSDD261 pKa = 3.95FHH263 pKa = 8.69ARR265 pKa = 11.84FPAGASIPAGGFQTVALAGSEE286 pKa = 4.31AFFAEE291 pKa = 4.72YY292 pKa = 10.2GVQPTYY298 pKa = 11.34EE299 pKa = 4.1LFEE302 pKa = 6.89DD303 pKa = 5.1GAADD307 pKa = 5.53AIPDD311 pKa = 3.66MRR313 pKa = 11.84EE314 pKa = 3.86ALPGSINAQGGLTNSGEE331 pKa = 4.53VVVLYY336 pKa = 10.36HH337 pKa = 6.7WDD339 pKa = 3.54EE340 pKa = 4.57SSDD343 pKa = 3.68LVADD347 pKa = 4.38LDD349 pKa = 4.13YY350 pKa = 11.4AVWGDD355 pKa = 3.27KK356 pKa = 11.17DD357 pKa = 3.7EE358 pKa = 5.72AVDD361 pKa = 3.68KK362 pKa = 11.11TGVALDD368 pKa = 4.58GPDD371 pKa = 3.57GDD373 pKa = 4.54TLVSEE378 pKa = 4.84YY379 pKa = 11.19AADD382 pKa = 3.94TPIASQDD389 pKa = 3.82VISGGAHH396 pKa = 6.41AFGEE400 pKa = 4.69SFTRR404 pKa = 11.84IDD406 pKa = 3.53FTEE409 pKa = 4.64GSEE412 pKa = 4.27TQSGGNGVNGADD424 pKa = 3.37EE425 pKa = 4.28TSEE428 pKa = 4.01NLSVTWASRR437 pKa = 11.84TPSPNSDD444 pKa = 3.88YY445 pKa = 11.31VPPTPDD451 pKa = 3.21LVITEE456 pKa = 4.12VMQNPAAVGDD466 pKa = 4.29SSGEE470 pKa = 3.59WFEE473 pKa = 3.7IHH475 pKa = 6.59NAGDD479 pKa = 3.31VDD481 pKa = 3.82VDD483 pKa = 3.68INGWTIADD491 pKa = 4.37LGSDD495 pKa = 3.4SHH497 pKa = 7.53VINAGGPLVVPAGGYY512 pKa = 8.05MVLGNNADD520 pKa = 3.69SGSNGGLVVDD530 pKa = 4.13YY531 pKa = 10.32AYY533 pKa = 11.23SGVFLANGDD542 pKa = 4.08DD543 pKa = 3.77EE544 pKa = 5.98LVLFDD549 pKa = 3.93AAGFEE554 pKa = 4.48VDD556 pKa = 3.55GVAWDD561 pKa = 4.85GGPVWPDD568 pKa = 3.01PTGASMALIDD578 pKa = 5.65LALDD582 pKa = 3.9NNDD585 pKa = 3.09GNNWCEE591 pKa = 4.0SQNPYY596 pKa = 11.1GDD598 pKa = 3.89GDD600 pKa = 3.74RR601 pKa = 11.84GTPGVANSCEE611 pKa = 3.9VLIPEE616 pKa = 5.44FGACGDD622 pKa = 3.84AATKK626 pKa = 9.92IHH628 pKa = 6.56AVQGNGLASPIAGSAGVIIEE648 pKa = 4.22GVVVGDD654 pKa = 3.5FQGGDD659 pKa = 3.14RR660 pKa = 11.84LNGFFVQEE668 pKa = 3.95EE669 pKa = 4.37SADD672 pKa = 3.74VDD674 pKa = 3.66GDD676 pKa = 3.86PATSEE681 pKa = 4.55GIFVYY686 pKa = 10.6DD687 pKa = 3.44EE688 pKa = 4.19NFGVDD693 pKa = 3.65VNVGDD698 pKa = 3.91VVRR701 pKa = 11.84VQGTVIEE708 pKa = 4.88FFDD711 pKa = 3.86LTEE714 pKa = 4.15ISPVINLVTCGTGTADD730 pKa = 3.53TQDD733 pKa = 3.14ILLPLAGPDD742 pKa = 3.24ALEE745 pKa = 4.24ALEE748 pKa = 4.35GMSVHH753 pKa = 7.03LPQTLFVTDD762 pKa = 3.68NFTLGRR768 pKa = 11.84YY769 pKa = 9.66GEE771 pKa = 4.25VGLAVGGPLDD781 pKa = 3.48IPTNVVAPGAAAQALQAQNDD801 pKa = 3.82LSRR804 pKa = 11.84IQLDD808 pKa = 4.14DD809 pKa = 3.85GSGLQNPVPVPPYY822 pKa = 9.31FAPDD826 pKa = 3.11GTLRR830 pKa = 11.84VGDD833 pKa = 3.73ATSAVTGVLGFGFGEE848 pKa = 4.4YY849 pKa = 9.7EE850 pKa = 3.74IHH852 pKa = 6.35PTDD855 pKa = 3.09VVAFSRR861 pKa = 11.84EE862 pKa = 3.79NEE864 pKa = 4.27RR865 pKa = 11.84PDD867 pKa = 4.1DD868 pKa = 4.45APDD871 pKa = 3.26VGNPLVTVAGFNVLNYY887 pKa = 7.55FTTLDD892 pKa = 4.43DD893 pKa = 3.81NTPVCGPSASMDD905 pKa = 3.22CRR907 pKa = 11.84GAEE910 pKa = 3.69NAFEE914 pKa = 4.44FEE916 pKa = 4.29RR917 pKa = 11.84QRR919 pKa = 11.84SKK921 pKa = 10.7IVAALSLLGADD932 pKa = 3.16VVGLVEE938 pKa = 4.27VEE940 pKa = 4.08NAAGDD945 pKa = 4.27GPVADD950 pKa = 5.03LVDD953 pKa = 5.33GINDD957 pKa = 3.22ILGADD962 pKa = 4.18TYY964 pKa = 11.71AYY966 pKa = 9.54IATGAIGTDD975 pKa = 4.44AIRR978 pKa = 11.84QALIYY983 pKa = 10.41KK984 pKa = 8.98PDD986 pKa = 3.28AVTPVGGFEE995 pKa = 4.46TLDD998 pKa = 3.77SSDD1001 pKa = 4.37DD1002 pKa = 3.57PAFIDD1007 pKa = 3.7TANRR1011 pKa = 11.84PALAQSFTDD1020 pKa = 4.19GSTGEE1025 pKa = 4.33VFTVAVNHH1033 pKa = 6.36LKK1035 pKa = 10.85SKK1037 pKa = 10.9GSDD1040 pKa = 3.4CDD1042 pKa = 3.77GLGDD1046 pKa = 4.84PDD1048 pKa = 4.01TGDD1051 pKa = 3.44GQGNCNLIRR1060 pKa = 11.84AAAASALATWLAADD1074 pKa = 3.8PTGSGSEE1081 pKa = 3.72RR1082 pKa = 11.84SLIIGDD1088 pKa = 4.15LNAYY1092 pKa = 9.83AKK1094 pKa = 9.95EE1095 pKa = 4.02DD1096 pKa = 4.57PIVVLKK1102 pKa = 10.93DD1103 pKa = 3.09AGFVDD1108 pKa = 5.13LVEE1111 pKa = 4.57VYY1113 pKa = 10.85NGVGYY1118 pKa = 11.02ADD1120 pKa = 3.66GAYY1123 pKa = 10.06SYY1125 pKa = 11.65GFGGQMGYY1133 pKa = 10.54LDD1135 pKa = 4.38HH1136 pKa = 7.3ALASAAFAADD1146 pKa = 3.5ASGATAWHH1154 pKa = 6.63INADD1158 pKa = 3.57EE1159 pKa = 4.33PAALDD1164 pKa = 4.14YY1165 pKa = 11.32NDD1167 pKa = 4.96HH1168 pKa = 5.99NQDD1171 pKa = 3.38VLFSPDD1177 pKa = 3.28PYY1179 pKa = 10.28RR1180 pKa = 11.84ASDD1183 pKa = 3.35HH1184 pKa = 6.7DD1185 pKa = 4.14AVVVGLFNDD1194 pKa = 3.71EE1195 pKa = 4.91DD1196 pKa = 5.36DD1197 pKa = 4.86DD1198 pKa = 5.1GVWDD1202 pKa = 5.21TIDD1205 pKa = 3.64ACPATVIPEE1214 pKa = 4.21TPPTRR1219 pKa = 11.84GLGNNRR1225 pKa = 11.84FALLDD1230 pKa = 4.1DD1231 pKa = 5.65DD1232 pKa = 5.75GVFDD1236 pKa = 3.88TTEE1239 pKa = 4.05GGGPGVSFTIHH1250 pKa = 6.38DD1251 pKa = 3.85TAGCSCEE1258 pKa = 4.12QIIAAADD1265 pKa = 3.88LGAGHH1270 pKa = 7.37AKK1272 pKa = 9.99FGCSIGEE1279 pKa = 3.74IRR1281 pKa = 11.84DD1282 pKa = 3.58WVSAQQ1287 pKa = 3.07
Molecular weight: 132.28 kDa
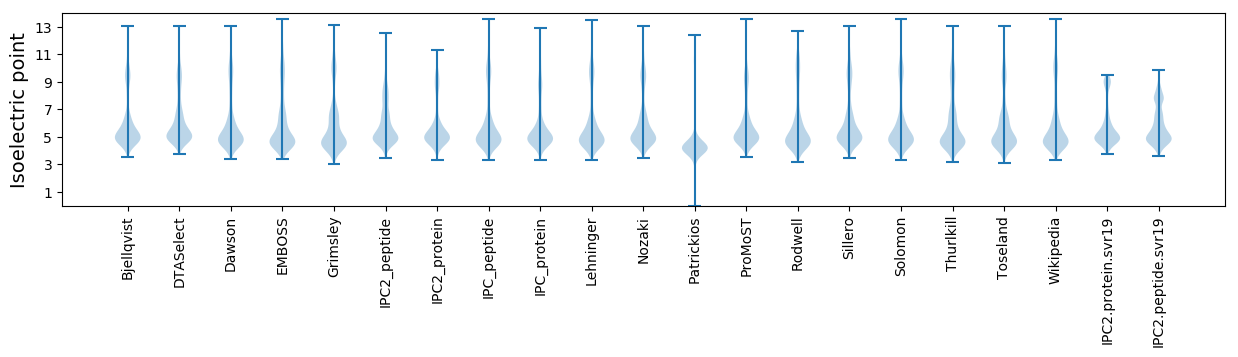
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5N0T971|A0A5N0T971_9GAMM DUF481 domain-containing protein OS=Wenzhouxiangella sp. W260 OX=2613842 GN=F3N42_13780 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.56RR3 pKa = 11.84TFQPSRR9 pKa = 11.84LKK11 pKa = 10.1RR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.41GFRR19 pKa = 11.84ARR21 pKa = 11.84MATRR25 pKa = 11.84GGRR28 pKa = 11.84AVINARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.67GRR39 pKa = 11.84KK40 pKa = 8.91RR41 pKa = 11.84LSAA44 pKa = 3.96
MM1 pKa = 7.45KK2 pKa = 9.56RR3 pKa = 11.84TFQPSRR9 pKa = 11.84LKK11 pKa = 10.1RR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.41GFRR19 pKa = 11.84ARR21 pKa = 11.84MATRR25 pKa = 11.84GGRR28 pKa = 11.84AVINARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.67GRR39 pKa = 11.84KK40 pKa = 8.91RR41 pKa = 11.84LSAA44 pKa = 3.96
Molecular weight: 5.14 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1066961 |
25 |
2914 |
350.7 |
38.3 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.541 ± 0.064 | 0.868 ± 0.017 |
6.664 ± 0.038 | 6.089 ± 0.04 |
3.676 ± 0.029 | 8.662 ± 0.036 |
2.153 ± 0.021 | 4.506 ± 0.032 |
2.485 ± 0.034 | 10.199 ± 0.052 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.434 ± 0.02 | 2.888 ± 0.026 |
5.108 ± 0.032 | 3.405 ± 0.025 |
7.048 ± 0.048 | 5.326 ± 0.034 |
5.23 ± 0.04 | 7.569 ± 0.036 |
1.643 ± 0.022 | 2.506 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
