
Reovirus type 3 (strain Dearing) (T3D) (Mammalian orthoreovirus 3)
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Resentoviricetes; Reovirales; Reoviridae; Spinareovirinae; Orthoreovirus; Mammalian orthoreovirus; Mammalian orthoreovirus 3
Average proteome isoelectric point is 5.79
Get precalculated fractions of proteins
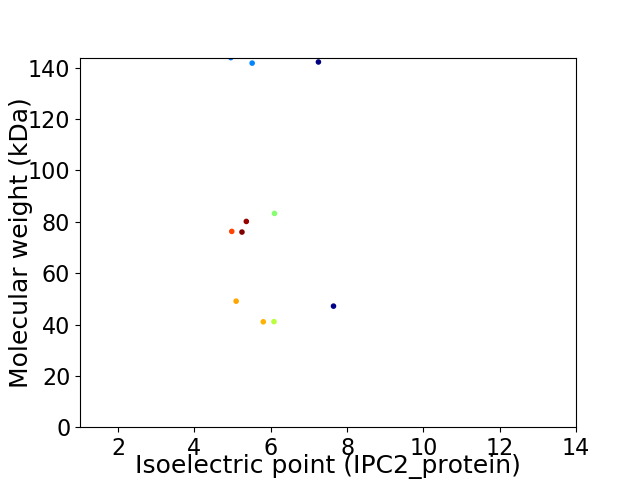
Virtual 2D-PAGE plot for 11 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
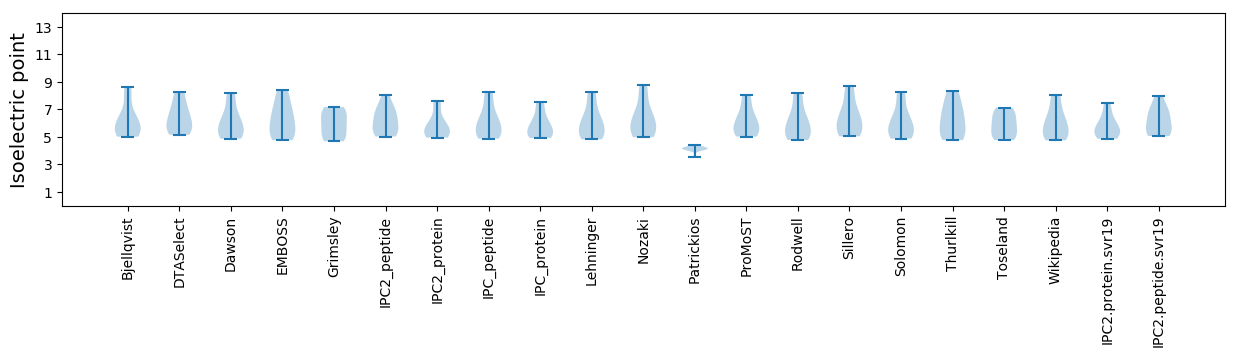
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>sp|P12418|MU2_REOVD Microtubule-associated protein mu-2 OS=Reovirus type 3 (strain Dearing) OX=10886 GN=M1 PE=1 SV=1
MM1 pKa = 7.34ANVWGVRR8 pKa = 11.84LADD11 pKa = 3.82SLSSPTIEE19 pKa = 4.01TRR21 pKa = 11.84TRR23 pKa = 11.84QYY25 pKa = 10.47TLHH28 pKa = 7.18DD29 pKa = 4.26LCSDD33 pKa = 3.92LDD35 pKa = 4.12ANPGRR40 pKa = 11.84EE41 pKa = 3.64PWKK44 pKa = 9.54PLRR47 pKa = 11.84NQRR50 pKa = 11.84TNNIVAVQLFRR61 pKa = 11.84PLQGLVLDD69 pKa = 4.01TQLYY73 pKa = 8.21GFPGAFDD80 pKa = 3.07DD81 pKa = 3.81WEE83 pKa = 4.65RR84 pKa = 11.84FMRR87 pKa = 11.84EE88 pKa = 3.38KK89 pKa = 11.01LRR91 pKa = 11.84VLKK94 pKa = 11.15YY95 pKa = 9.5EE96 pKa = 3.91VLRR99 pKa = 11.84IYY101 pKa = 10.23PISNYY106 pKa = 10.13SNEE109 pKa = 4.08HH110 pKa = 5.13VNVFVANALVGAFLSNQAFYY130 pKa = 11.48DD131 pKa = 4.14LLPLLIINDD140 pKa = 3.8TMIGDD145 pKa = 4.58LLGTGASLSQFFQSHH160 pKa = 6.04GDD162 pKa = 3.68VLEE165 pKa = 4.24VAAGRR170 pKa = 11.84KK171 pKa = 7.8YY172 pKa = 10.94LQMEE176 pKa = 4.56NYY178 pKa = 10.61SNDD181 pKa = 3.83DD182 pKa = 3.58DD183 pKa = 5.86DD184 pKa = 5.56PPLFAKK190 pKa = 10.55DD191 pKa = 3.38LSDD194 pKa = 3.66YY195 pKa = 11.6AKK197 pKa = 10.8AFYY200 pKa = 10.69SDD202 pKa = 3.4TYY204 pKa = 10.92EE205 pKa = 3.91VLDD208 pKa = 3.77RR209 pKa = 11.84FFWTHH214 pKa = 6.75DD215 pKa = 3.09SSAGVLVHH223 pKa = 6.38YY224 pKa = 8.23DD225 pKa = 3.06KK226 pKa = 10.18PTNGHH231 pKa = 6.61HH232 pKa = 6.2YY233 pKa = 11.06LLGTLTQMVSAPPYY247 pKa = 10.06IINATDD253 pKa = 3.2AMLLEE258 pKa = 4.74SCLEE262 pKa = 3.89QFSANVRR269 pKa = 11.84ARR271 pKa = 11.84PAQPVTRR278 pKa = 11.84LDD280 pKa = 3.2QCYY283 pKa = 9.51HH284 pKa = 5.99LRR286 pKa = 11.84WGAQYY291 pKa = 10.87VGEE294 pKa = 4.72DD295 pKa = 3.3SLTYY299 pKa = 10.4RR300 pKa = 11.84LGVLSLLATNGYY312 pKa = 9.17QLARR316 pKa = 11.84PIPRR320 pKa = 11.84QLTNRR325 pKa = 11.84WLSSFVSQIMSDD337 pKa = 3.81GVNEE341 pKa = 4.18TPLWPQEE348 pKa = 4.03RR349 pKa = 11.84YY350 pKa = 8.46VQIAYY355 pKa = 9.43DD356 pKa = 3.79SPSVVDD362 pKa = 4.59GATQYY367 pKa = 11.67GYY369 pKa = 10.63VRR371 pKa = 11.84KK372 pKa = 9.02NQLRR376 pKa = 11.84LGMRR380 pKa = 11.84ISALQSLSDD389 pKa = 3.84TPSPVQWLPQYY400 pKa = 10.55TIDD403 pKa = 4.28QAAMDD408 pKa = 4.7EE409 pKa = 4.25GDD411 pKa = 4.05LMVSRR416 pKa = 11.84LTQLPLRR423 pKa = 11.84PDD425 pKa = 3.52YY426 pKa = 11.63GNIWVGDD433 pKa = 3.82ALSYY437 pKa = 10.3YY438 pKa = 10.06VDD440 pKa = 3.67YY441 pKa = 11.11NRR443 pKa = 11.84SHH445 pKa = 6.66RR446 pKa = 11.84VVLSSEE452 pKa = 4.47LPQLPDD458 pKa = 2.96TYY460 pKa = 11.3FDD462 pKa = 3.83GDD464 pKa = 3.65EE465 pKa = 4.0QYY467 pKa = 11.5GRR469 pKa = 11.84SLFSLARR476 pKa = 11.84KK477 pKa = 9.61IGDD480 pKa = 3.21RR481 pKa = 11.84SLVKK485 pKa = 9.91DD486 pKa = 3.47TAVLKK491 pKa = 10.42HH492 pKa = 6.56AYY494 pKa = 8.89QAIDD498 pKa = 3.52PNTGKK503 pKa = 10.6EE504 pKa = 4.1YY505 pKa = 10.63LRR507 pKa = 11.84SRR509 pKa = 11.84QSVAYY514 pKa = 8.95FGASAGHH521 pKa = 6.32SGADD525 pKa = 3.12QPLVIEE531 pKa = 4.54PWIQGKK537 pKa = 9.89ISGVPPPSSVRR548 pKa = 11.84QFGYY552 pKa = 10.76DD553 pKa = 3.29VARR556 pKa = 11.84GAIVDD561 pKa = 4.35LARR564 pKa = 11.84PFPSGDD570 pKa = 3.41YY571 pKa = 10.22QFVYY575 pKa = 10.9SDD577 pKa = 3.15VDD579 pKa = 3.69QVVDD583 pKa = 3.69GHH585 pKa = 7.25DD586 pKa = 4.11DD587 pKa = 3.62LSISSGLVEE596 pKa = 4.94SLLSSCMHH604 pKa = 6.35ATAPGGSFVVKK615 pKa = 10.23INFPTRR621 pKa = 11.84PVWHH625 pKa = 6.69YY626 pKa = 10.51IEE628 pKa = 5.3QKK630 pKa = 9.72ILPNITSYY638 pKa = 10.97MLIKK642 pKa = 10.53PFVTNNVEE650 pKa = 4.06LFFVAFGVHH659 pKa = 4.71QHH661 pKa = 6.6SSLTWTSGVYY671 pKa = 10.02FFLVDD676 pKa = 3.09HH677 pKa = 7.04FYY679 pKa = 10.67RR680 pKa = 11.84YY681 pKa = 7.46EE682 pKa = 3.98TLSTISRR689 pKa = 11.84QLPSFGYY696 pKa = 10.34VDD698 pKa = 4.85DD699 pKa = 5.1GSSVTGIEE707 pKa = 4.39TISIEE712 pKa = 4.16NPGFSNMTQAARR724 pKa = 11.84IGISGLCANVGNARR738 pKa = 11.84KK739 pKa = 9.98SIAIYY744 pKa = 8.9EE745 pKa = 4.3SHH747 pKa = 6.82GARR750 pKa = 11.84VLTITSRR757 pKa = 11.84RR758 pKa = 11.84SPASARR764 pKa = 11.84RR765 pKa = 11.84KK766 pKa = 9.65SRR768 pKa = 11.84LRR770 pKa = 11.84YY771 pKa = 9.57LPLIDD776 pKa = 4.18PRR778 pKa = 11.84SLEE781 pKa = 4.04VQARR785 pKa = 11.84TILPADD791 pKa = 3.38PVLFEE796 pKa = 4.41NVSGASPHH804 pKa = 5.75VCLTMMYY811 pKa = 9.72NFEE814 pKa = 4.24VSSAVYY820 pKa = 10.61DD821 pKa = 3.73GDD823 pKa = 3.94VVLDD827 pKa = 4.5LGTGPEE833 pKa = 3.96AKK835 pKa = 9.76ILEE838 pKa = 5.29LIPATSPVTCVDD850 pKa = 3.28IRR852 pKa = 11.84PTAQPSGCWNVRR864 pKa = 11.84TTFLEE869 pKa = 3.91LDD871 pKa = 3.83YY872 pKa = 11.51LSDD875 pKa = 3.26GWITGVRR882 pKa = 11.84GDD884 pKa = 4.46IVTCMLSLGAAAAGKK899 pKa = 10.91SMTFDD904 pKa = 3.32AAFQQLIKK912 pKa = 10.78VLSKK916 pKa = 9.66STANVVLVQVNCPTDD931 pKa = 3.4VVRR934 pKa = 11.84SIKK937 pKa = 10.54GYY939 pKa = 10.91LEE941 pKa = 3.89IDD943 pKa = 3.83STNKK947 pKa = 10.08RR948 pKa = 11.84YY949 pKa = 10.15RR950 pKa = 11.84FPKK953 pKa = 10.01FGRR956 pKa = 11.84DD957 pKa = 3.29EE958 pKa = 4.96PYY960 pKa = 10.91SDD962 pKa = 3.58MDD964 pKa = 4.02ALEE967 pKa = 4.72KK968 pKa = 10.16ICRR971 pKa = 11.84TAWPNCSITWVPLSYY986 pKa = 10.89DD987 pKa = 3.71LRR989 pKa = 11.84WTRR992 pKa = 11.84LALLEE997 pKa = 4.23STTLSSASIRR1007 pKa = 11.84IAEE1010 pKa = 4.22LMYY1013 pKa = 10.6KK1014 pKa = 9.97YY1015 pKa = 9.76MPIMRR1020 pKa = 11.84IDD1022 pKa = 3.18IHH1024 pKa = 6.13GLPMEE1029 pKa = 4.29KK1030 pKa = 9.6RR1031 pKa = 11.84GNFIVGQNCSLVIPGFNAQDD1051 pKa = 3.46VFNCYY1056 pKa = 9.73FNSALAFSTEE1066 pKa = 4.13DD1067 pKa = 3.22VNAAMIPQVSAQFDD1081 pKa = 3.63ATKK1084 pKa = 11.06GEE1086 pKa = 4.08WTLDD1090 pKa = 3.42MVFSDD1095 pKa = 3.23AGIYY1099 pKa = 8.99TMQALVGSNANPVSLGSFVVDD1120 pKa = 4.37SPDD1123 pKa = 4.22VDD1125 pKa = 3.82ITDD1128 pKa = 3.6AWPAQLDD1135 pKa = 3.67FTIAGTDD1142 pKa = 2.91VDD1144 pKa = 3.83ITVNPYY1150 pKa = 8.82YY1151 pKa = 10.96RR1152 pKa = 11.84LMTFVRR1158 pKa = 11.84IDD1160 pKa = 3.81GQWQIANPDD1169 pKa = 3.1KK1170 pKa = 11.12FQFFSSASGTLVMNVKK1186 pKa = 10.45LDD1188 pKa = 3.41IADD1191 pKa = 4.19KK1192 pKa = 10.68YY1193 pKa = 10.02LLYY1196 pKa = 10.57YY1197 pKa = 9.88IRR1199 pKa = 11.84DD1200 pKa = 3.64VQSRR1204 pKa = 11.84DD1205 pKa = 3.04VGFYY1209 pKa = 9.9IQHH1212 pKa = 7.14PLQLLNTITLPTNEE1226 pKa = 5.26DD1227 pKa = 3.81LFLSAPDD1234 pKa = 3.44MRR1236 pKa = 11.84EE1237 pKa = 3.61WAVKK1241 pKa = 10.18EE1242 pKa = 4.15SGNTICILNSQGFVLPQDD1260 pKa = 3.21WDD1262 pKa = 3.8VLTDD1266 pKa = 4.23TISWSPSIPTYY1277 pKa = 10.24IVPPGDD1283 pKa = 3.52YY1284 pKa = 10.21TLTPLL1289 pKa = 4.79
MM1 pKa = 7.34ANVWGVRR8 pKa = 11.84LADD11 pKa = 3.82SLSSPTIEE19 pKa = 4.01TRR21 pKa = 11.84TRR23 pKa = 11.84QYY25 pKa = 10.47TLHH28 pKa = 7.18DD29 pKa = 4.26LCSDD33 pKa = 3.92LDD35 pKa = 4.12ANPGRR40 pKa = 11.84EE41 pKa = 3.64PWKK44 pKa = 9.54PLRR47 pKa = 11.84NQRR50 pKa = 11.84TNNIVAVQLFRR61 pKa = 11.84PLQGLVLDD69 pKa = 4.01TQLYY73 pKa = 8.21GFPGAFDD80 pKa = 3.07DD81 pKa = 3.81WEE83 pKa = 4.65RR84 pKa = 11.84FMRR87 pKa = 11.84EE88 pKa = 3.38KK89 pKa = 11.01LRR91 pKa = 11.84VLKK94 pKa = 11.15YY95 pKa = 9.5EE96 pKa = 3.91VLRR99 pKa = 11.84IYY101 pKa = 10.23PISNYY106 pKa = 10.13SNEE109 pKa = 4.08HH110 pKa = 5.13VNVFVANALVGAFLSNQAFYY130 pKa = 11.48DD131 pKa = 4.14LLPLLIINDD140 pKa = 3.8TMIGDD145 pKa = 4.58LLGTGASLSQFFQSHH160 pKa = 6.04GDD162 pKa = 3.68VLEE165 pKa = 4.24VAAGRR170 pKa = 11.84KK171 pKa = 7.8YY172 pKa = 10.94LQMEE176 pKa = 4.56NYY178 pKa = 10.61SNDD181 pKa = 3.83DD182 pKa = 3.58DD183 pKa = 5.86DD184 pKa = 5.56PPLFAKK190 pKa = 10.55DD191 pKa = 3.38LSDD194 pKa = 3.66YY195 pKa = 11.6AKK197 pKa = 10.8AFYY200 pKa = 10.69SDD202 pKa = 3.4TYY204 pKa = 10.92EE205 pKa = 3.91VLDD208 pKa = 3.77RR209 pKa = 11.84FFWTHH214 pKa = 6.75DD215 pKa = 3.09SSAGVLVHH223 pKa = 6.38YY224 pKa = 8.23DD225 pKa = 3.06KK226 pKa = 10.18PTNGHH231 pKa = 6.61HH232 pKa = 6.2YY233 pKa = 11.06LLGTLTQMVSAPPYY247 pKa = 10.06IINATDD253 pKa = 3.2AMLLEE258 pKa = 4.74SCLEE262 pKa = 3.89QFSANVRR269 pKa = 11.84ARR271 pKa = 11.84PAQPVTRR278 pKa = 11.84LDD280 pKa = 3.2QCYY283 pKa = 9.51HH284 pKa = 5.99LRR286 pKa = 11.84WGAQYY291 pKa = 10.87VGEE294 pKa = 4.72DD295 pKa = 3.3SLTYY299 pKa = 10.4RR300 pKa = 11.84LGVLSLLATNGYY312 pKa = 9.17QLARR316 pKa = 11.84PIPRR320 pKa = 11.84QLTNRR325 pKa = 11.84WLSSFVSQIMSDD337 pKa = 3.81GVNEE341 pKa = 4.18TPLWPQEE348 pKa = 4.03RR349 pKa = 11.84YY350 pKa = 8.46VQIAYY355 pKa = 9.43DD356 pKa = 3.79SPSVVDD362 pKa = 4.59GATQYY367 pKa = 11.67GYY369 pKa = 10.63VRR371 pKa = 11.84KK372 pKa = 9.02NQLRR376 pKa = 11.84LGMRR380 pKa = 11.84ISALQSLSDD389 pKa = 3.84TPSPVQWLPQYY400 pKa = 10.55TIDD403 pKa = 4.28QAAMDD408 pKa = 4.7EE409 pKa = 4.25GDD411 pKa = 4.05LMVSRR416 pKa = 11.84LTQLPLRR423 pKa = 11.84PDD425 pKa = 3.52YY426 pKa = 11.63GNIWVGDD433 pKa = 3.82ALSYY437 pKa = 10.3YY438 pKa = 10.06VDD440 pKa = 3.67YY441 pKa = 11.11NRR443 pKa = 11.84SHH445 pKa = 6.66RR446 pKa = 11.84VVLSSEE452 pKa = 4.47LPQLPDD458 pKa = 2.96TYY460 pKa = 11.3FDD462 pKa = 3.83GDD464 pKa = 3.65EE465 pKa = 4.0QYY467 pKa = 11.5GRR469 pKa = 11.84SLFSLARR476 pKa = 11.84KK477 pKa = 9.61IGDD480 pKa = 3.21RR481 pKa = 11.84SLVKK485 pKa = 9.91DD486 pKa = 3.47TAVLKK491 pKa = 10.42HH492 pKa = 6.56AYY494 pKa = 8.89QAIDD498 pKa = 3.52PNTGKK503 pKa = 10.6EE504 pKa = 4.1YY505 pKa = 10.63LRR507 pKa = 11.84SRR509 pKa = 11.84QSVAYY514 pKa = 8.95FGASAGHH521 pKa = 6.32SGADD525 pKa = 3.12QPLVIEE531 pKa = 4.54PWIQGKK537 pKa = 9.89ISGVPPPSSVRR548 pKa = 11.84QFGYY552 pKa = 10.76DD553 pKa = 3.29VARR556 pKa = 11.84GAIVDD561 pKa = 4.35LARR564 pKa = 11.84PFPSGDD570 pKa = 3.41YY571 pKa = 10.22QFVYY575 pKa = 10.9SDD577 pKa = 3.15VDD579 pKa = 3.69QVVDD583 pKa = 3.69GHH585 pKa = 7.25DD586 pKa = 4.11DD587 pKa = 3.62LSISSGLVEE596 pKa = 4.94SLLSSCMHH604 pKa = 6.35ATAPGGSFVVKK615 pKa = 10.23INFPTRR621 pKa = 11.84PVWHH625 pKa = 6.69YY626 pKa = 10.51IEE628 pKa = 5.3QKK630 pKa = 9.72ILPNITSYY638 pKa = 10.97MLIKK642 pKa = 10.53PFVTNNVEE650 pKa = 4.06LFFVAFGVHH659 pKa = 4.71QHH661 pKa = 6.6SSLTWTSGVYY671 pKa = 10.02FFLVDD676 pKa = 3.09HH677 pKa = 7.04FYY679 pKa = 10.67RR680 pKa = 11.84YY681 pKa = 7.46EE682 pKa = 3.98TLSTISRR689 pKa = 11.84QLPSFGYY696 pKa = 10.34VDD698 pKa = 4.85DD699 pKa = 5.1GSSVTGIEE707 pKa = 4.39TISIEE712 pKa = 4.16NPGFSNMTQAARR724 pKa = 11.84IGISGLCANVGNARR738 pKa = 11.84KK739 pKa = 9.98SIAIYY744 pKa = 8.9EE745 pKa = 4.3SHH747 pKa = 6.82GARR750 pKa = 11.84VLTITSRR757 pKa = 11.84RR758 pKa = 11.84SPASARR764 pKa = 11.84RR765 pKa = 11.84KK766 pKa = 9.65SRR768 pKa = 11.84LRR770 pKa = 11.84YY771 pKa = 9.57LPLIDD776 pKa = 4.18PRR778 pKa = 11.84SLEE781 pKa = 4.04VQARR785 pKa = 11.84TILPADD791 pKa = 3.38PVLFEE796 pKa = 4.41NVSGASPHH804 pKa = 5.75VCLTMMYY811 pKa = 9.72NFEE814 pKa = 4.24VSSAVYY820 pKa = 10.61DD821 pKa = 3.73GDD823 pKa = 3.94VVLDD827 pKa = 4.5LGTGPEE833 pKa = 3.96AKK835 pKa = 9.76ILEE838 pKa = 5.29LIPATSPVTCVDD850 pKa = 3.28IRR852 pKa = 11.84PTAQPSGCWNVRR864 pKa = 11.84TTFLEE869 pKa = 3.91LDD871 pKa = 3.83YY872 pKa = 11.51LSDD875 pKa = 3.26GWITGVRR882 pKa = 11.84GDD884 pKa = 4.46IVTCMLSLGAAAAGKK899 pKa = 10.91SMTFDD904 pKa = 3.32AAFQQLIKK912 pKa = 10.78VLSKK916 pKa = 9.66STANVVLVQVNCPTDD931 pKa = 3.4VVRR934 pKa = 11.84SIKK937 pKa = 10.54GYY939 pKa = 10.91LEE941 pKa = 3.89IDD943 pKa = 3.83STNKK947 pKa = 10.08RR948 pKa = 11.84YY949 pKa = 10.15RR950 pKa = 11.84FPKK953 pKa = 10.01FGRR956 pKa = 11.84DD957 pKa = 3.29EE958 pKa = 4.96PYY960 pKa = 10.91SDD962 pKa = 3.58MDD964 pKa = 4.02ALEE967 pKa = 4.72KK968 pKa = 10.16ICRR971 pKa = 11.84TAWPNCSITWVPLSYY986 pKa = 10.89DD987 pKa = 3.71LRR989 pKa = 11.84WTRR992 pKa = 11.84LALLEE997 pKa = 4.23STTLSSASIRR1007 pKa = 11.84IAEE1010 pKa = 4.22LMYY1013 pKa = 10.6KK1014 pKa = 9.97YY1015 pKa = 9.76MPIMRR1020 pKa = 11.84IDD1022 pKa = 3.18IHH1024 pKa = 6.13GLPMEE1029 pKa = 4.29KK1030 pKa = 9.6RR1031 pKa = 11.84GNFIVGQNCSLVIPGFNAQDD1051 pKa = 3.46VFNCYY1056 pKa = 9.73FNSALAFSTEE1066 pKa = 4.13DD1067 pKa = 3.22VNAAMIPQVSAQFDD1081 pKa = 3.63ATKK1084 pKa = 11.06GEE1086 pKa = 4.08WTLDD1090 pKa = 3.42MVFSDD1095 pKa = 3.23AGIYY1099 pKa = 8.99TMQALVGSNANPVSLGSFVVDD1120 pKa = 4.37SPDD1123 pKa = 4.22VDD1125 pKa = 3.82ITDD1128 pKa = 3.6AWPAQLDD1135 pKa = 3.67FTIAGTDD1142 pKa = 2.91VDD1144 pKa = 3.83ITVNPYY1150 pKa = 8.82YY1151 pKa = 10.96RR1152 pKa = 11.84LMTFVRR1158 pKa = 11.84IDD1160 pKa = 3.81GQWQIANPDD1169 pKa = 3.1KK1170 pKa = 11.12FQFFSSASGTLVMNVKK1186 pKa = 10.45LDD1188 pKa = 3.41IADD1191 pKa = 4.19KK1192 pKa = 10.68YY1193 pKa = 10.02LLYY1196 pKa = 10.57YY1197 pKa = 9.88IRR1199 pKa = 11.84DD1200 pKa = 3.64VQSRR1204 pKa = 11.84DD1205 pKa = 3.04VGFYY1209 pKa = 9.9IQHH1212 pKa = 7.14PLQLLNTITLPTNEE1226 pKa = 5.26DD1227 pKa = 3.81LFLSAPDD1234 pKa = 3.44MRR1236 pKa = 11.84EE1237 pKa = 3.61WAVKK1241 pKa = 10.18EE1242 pKa = 4.15SGNTICILNSQGFVLPQDD1260 pKa = 3.21WDD1262 pKa = 3.8VLTDD1266 pKa = 4.23TISWSPSIPTYY1277 pKa = 10.24IVPPGDD1283 pKa = 3.52YY1284 pKa = 10.21TLTPLL1289 pKa = 4.79
Molecular weight: 143.98 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|P03526|SIGNS_REOVD Protein sigma-NS OS=Reovirus type 3 (strain Dearing) OX=10886 GN=S3 PE=2 SV=1
MM1 pKa = 7.31ARR3 pKa = 11.84AAFLFKK9 pKa = 10.33TVGFGGLQNVPINDD23 pKa = 3.83EE24 pKa = 4.26LSSHH28 pKa = 6.8LLRR31 pKa = 11.84AGNSPWQLTQFLDD44 pKa = 4.17WISLGRR50 pKa = 11.84GLATSALVPTAGSRR64 pKa = 11.84YY65 pKa = 8.74YY66 pKa = 11.13QMSCLLSGTLQIPFRR81 pKa = 11.84PNHH84 pKa = 5.6RR85 pKa = 11.84WGDD88 pKa = 3.35IRR90 pKa = 11.84FLRR93 pKa = 11.84LVWSAPTLDD102 pKa = 4.12GLVVAPPQVLAQPALQAQADD122 pKa = 4.12RR123 pKa = 11.84VYY125 pKa = 11.29DD126 pKa = 3.87CDD128 pKa = 5.2DD129 pKa = 3.71YY130 pKa = 11.78PFLARR135 pKa = 11.84DD136 pKa = 3.34PRR138 pKa = 11.84FKK140 pKa = 10.89HH141 pKa = 6.07RR142 pKa = 11.84VYY144 pKa = 10.5QQLSAVTLLNLTGFGPISYY163 pKa = 10.03VRR165 pKa = 11.84VDD167 pKa = 3.06EE168 pKa = 5.2DD169 pKa = 3.21MWSGDD174 pKa = 3.63VNQLLMNYY182 pKa = 9.49FGHH185 pKa = 6.38TFAEE189 pKa = 4.2IAYY192 pKa = 6.69TLCQASANRR201 pKa = 11.84PWEE204 pKa = 3.74YY205 pKa = 10.93DD206 pKa = 2.89GTYY209 pKa = 11.24ARR211 pKa = 11.84MTQIVLSLFWLSYY224 pKa = 10.67VGVIHH229 pKa = 6.07QQNTYY234 pKa = 8.17RR235 pKa = 11.84TFYY238 pKa = 9.35FQCNRR243 pKa = 11.84RR244 pKa = 11.84GDD246 pKa = 3.7AAEE249 pKa = 4.12VWILSCSLNHH259 pKa = 6.59SAQIRR264 pKa = 11.84PGNRR268 pKa = 11.84SLFVMPTSPDD278 pKa = 2.57WNMDD282 pKa = 3.01VNLILSSTLTGCLCSGSQLPLIDD305 pKa = 4.33NNSVPAVSRR314 pKa = 11.84NIHH317 pKa = 5.35GWTGRR322 pKa = 11.84AGNQLHH328 pKa = 6.48GFQVRR333 pKa = 11.84RR334 pKa = 11.84MVTEE338 pKa = 4.08FCDD341 pKa = 3.06RR342 pKa = 11.84LRR344 pKa = 11.84RR345 pKa = 11.84DD346 pKa = 3.17GVMTQAQQNQVEE358 pKa = 4.24ALADD362 pKa = 3.24QTQQFKK368 pKa = 10.57RR369 pKa = 11.84DD370 pKa = 3.67KK371 pKa = 10.52LEE373 pKa = 3.53TWARR377 pKa = 11.84EE378 pKa = 3.64DD379 pKa = 3.39DD380 pKa = 4.48QYY382 pKa = 12.13NQAHH386 pKa = 6.94PNSTMFRR393 pKa = 11.84TKK395 pKa = 10.55PFTNAQWGRR404 pKa = 11.84GNTGATSAAIAALII418 pKa = 4.13
MM1 pKa = 7.31ARR3 pKa = 11.84AAFLFKK9 pKa = 10.33TVGFGGLQNVPINDD23 pKa = 3.83EE24 pKa = 4.26LSSHH28 pKa = 6.8LLRR31 pKa = 11.84AGNSPWQLTQFLDD44 pKa = 4.17WISLGRR50 pKa = 11.84GLATSALVPTAGSRR64 pKa = 11.84YY65 pKa = 8.74YY66 pKa = 11.13QMSCLLSGTLQIPFRR81 pKa = 11.84PNHH84 pKa = 5.6RR85 pKa = 11.84WGDD88 pKa = 3.35IRR90 pKa = 11.84FLRR93 pKa = 11.84LVWSAPTLDD102 pKa = 4.12GLVVAPPQVLAQPALQAQADD122 pKa = 4.12RR123 pKa = 11.84VYY125 pKa = 11.29DD126 pKa = 3.87CDD128 pKa = 5.2DD129 pKa = 3.71YY130 pKa = 11.78PFLARR135 pKa = 11.84DD136 pKa = 3.34PRR138 pKa = 11.84FKK140 pKa = 10.89HH141 pKa = 6.07RR142 pKa = 11.84VYY144 pKa = 10.5QQLSAVTLLNLTGFGPISYY163 pKa = 10.03VRR165 pKa = 11.84VDD167 pKa = 3.06EE168 pKa = 5.2DD169 pKa = 3.21MWSGDD174 pKa = 3.63VNQLLMNYY182 pKa = 9.49FGHH185 pKa = 6.38TFAEE189 pKa = 4.2IAYY192 pKa = 6.69TLCQASANRR201 pKa = 11.84PWEE204 pKa = 3.74YY205 pKa = 10.93DD206 pKa = 2.89GTYY209 pKa = 11.24ARR211 pKa = 11.84MTQIVLSLFWLSYY224 pKa = 10.67VGVIHH229 pKa = 6.07QQNTYY234 pKa = 8.17RR235 pKa = 11.84TFYY238 pKa = 9.35FQCNRR243 pKa = 11.84RR244 pKa = 11.84GDD246 pKa = 3.7AAEE249 pKa = 4.12VWILSCSLNHH259 pKa = 6.59SAQIRR264 pKa = 11.84PGNRR268 pKa = 11.84SLFVMPTSPDD278 pKa = 2.57WNMDD282 pKa = 3.01VNLILSSTLTGCLCSGSQLPLIDD305 pKa = 4.33NNSVPAVSRR314 pKa = 11.84NIHH317 pKa = 5.35GWTGRR322 pKa = 11.84AGNQLHH328 pKa = 6.48GFQVRR333 pKa = 11.84RR334 pKa = 11.84MVTEE338 pKa = 4.08FCDD341 pKa = 3.06RR342 pKa = 11.84LRR344 pKa = 11.84RR345 pKa = 11.84DD346 pKa = 3.17GVMTQAQQNQVEE358 pKa = 4.24ALADD362 pKa = 3.24QTQQFKK368 pKa = 10.57RR369 pKa = 11.84DD370 pKa = 3.67KK371 pKa = 10.52LEE373 pKa = 3.53TWARR377 pKa = 11.84EE378 pKa = 3.64DD379 pKa = 3.39DD380 pKa = 4.48QYY382 pKa = 12.13NQAHH386 pKa = 6.94PNSTMFRR393 pKa = 11.84TKK395 pKa = 10.55PFTNAQWGRR404 pKa = 11.84GNTGATSAAIAALII418 pKa = 4.13
Molecular weight: 47.16 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
8281 |
365 |
1289 |
752.8 |
83.84 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.006 ± 0.375 | 1.352 ± 0.152 |
6.461 ± 0.296 | 4.371 ± 0.292 |
3.502 ± 0.177 | 5.482 ± 0.369 |
2.005 ± 0.222 | 5.664 ± 0.219 |
3.768 ± 0.36 | 9.576 ± 0.353 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.236 ± 0.282 | 3.997 ± 0.363 |
5.482 ± 0.41 | 4.685 ± 0.347 |
5.651 ± 0.242 | 8.369 ± 0.304 |
6.135 ± 0.407 | 7.366 ± 0.354 |
1.473 ± 0.176 | 3.417 ± 0.328 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
