
Valsa sordida
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; sordariomyceta; Sordariomycetes; Sordariomycetidae; Diaporthales; Valsaceae; Valsa
Average proteome isoelectric point is 6.28
Get precalculated fractions of proteins
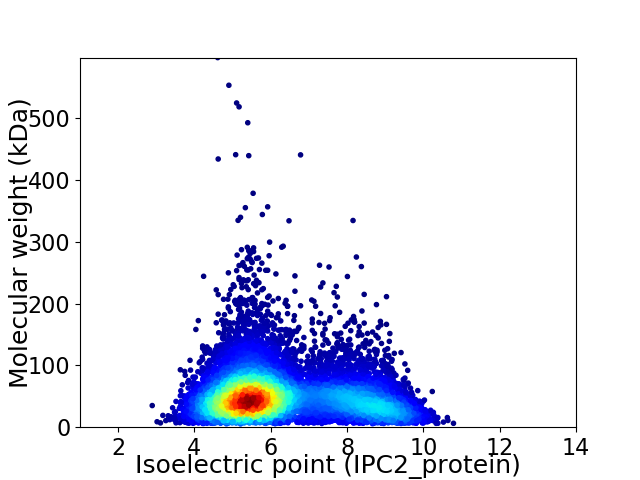
Virtual 2D-PAGE plot for 9818 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A423WCN8|A0A423WCN8_9PEZI Uncharacterized protein OS=Valsa sordida OX=252740 GN=VSDG_02845 PE=4 SV=1
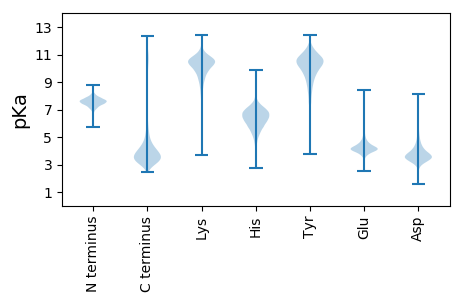
MM1 pKa = 7.96PSTITTAGVLAFMSLSGALAATNCTFSGSSGFSEE35 pKa = 4.42ISAKK39 pKa = 10.33KK40 pKa = 9.89SSCSTIIIDD49 pKa = 4.18SLEE52 pKa = 4.22VPAGEE57 pKa = 4.2TLDD60 pKa = 5.86LEE62 pKa = 4.57DD63 pKa = 5.93LNDD66 pKa = 3.74FTTLTSNKK74 pKa = 9.82VIFQGQTTWAYY85 pKa = 10.59AEE87 pKa = 4.34WEE89 pKa = 4.42GSLISISGNNITVKK103 pKa = 10.92GEE105 pKa = 3.95AGSALDD111 pKa = 4.03GQGALWWDD119 pKa = 3.5GLGGNGGVTKK129 pKa = 10.56PKK131 pKa = 10.16FFKK134 pKa = 10.81ANKK137 pKa = 9.96LYY139 pKa = 10.95DD140 pKa = 3.76SVLDD144 pKa = 5.1GITILNAPKK153 pKa = 10.49NSFSVNYY160 pKa = 10.06VEE162 pKa = 6.1NLVLQNIVINNTAGDD177 pKa = 4.08EE178 pKa = 4.19LNSDD182 pKa = 4.34GKK184 pKa = 8.85TLGHH188 pKa = 6.01NTDD191 pKa = 4.18AFDD194 pKa = 4.58INNCDD199 pKa = 2.99GVLFQNITVYY209 pKa = 10.99NQDD212 pKa = 3.01DD213 pKa = 4.16CVAVNSGEE221 pKa = 4.02NVVFRR226 pKa = 11.84DD227 pKa = 3.78ALCSGGHH234 pKa = 6.67GISIGSVGGRR244 pKa = 11.84SNNVVNNITFDD255 pKa = 3.54NVLMQNSQQSVRR267 pKa = 11.84IKK269 pKa = 10.09TIADD273 pKa = 3.51TNGTVSNVTYY283 pKa = 10.49RR284 pKa = 11.84NIVIDD289 pKa = 4.22SPTDD293 pKa = 3.25NTDD296 pKa = 2.85YY297 pKa = 11.44GIIVSQSYY305 pKa = 10.77NGVDD309 pKa = 3.62GEE311 pKa = 4.39PTNGVIISNFVLQNVTGTVYY331 pKa = 10.88EE332 pKa = 4.19DD333 pKa = 4.61AINIYY338 pKa = 9.48IEE340 pKa = 4.62CGEE343 pKa = 4.52GSCIDD348 pKa = 3.69WTWTDD353 pKa = 3.49VNVSGGKK360 pKa = 10.1DD361 pKa = 3.0SADD364 pKa = 3.54CMNIPDD370 pKa = 5.8GISCC374 pKa = 4.99
MM1 pKa = 7.96PSTITTAGVLAFMSLSGALAATNCTFSGSSGFSEE35 pKa = 4.42ISAKK39 pKa = 10.33KK40 pKa = 9.89SSCSTIIIDD49 pKa = 4.18SLEE52 pKa = 4.22VPAGEE57 pKa = 4.2TLDD60 pKa = 5.86LEE62 pKa = 4.57DD63 pKa = 5.93LNDD66 pKa = 3.74FTTLTSNKK74 pKa = 9.82VIFQGQTTWAYY85 pKa = 10.59AEE87 pKa = 4.34WEE89 pKa = 4.42GSLISISGNNITVKK103 pKa = 10.92GEE105 pKa = 3.95AGSALDD111 pKa = 4.03GQGALWWDD119 pKa = 3.5GLGGNGGVTKK129 pKa = 10.56PKK131 pKa = 10.16FFKK134 pKa = 10.81ANKK137 pKa = 9.96LYY139 pKa = 10.95DD140 pKa = 3.76SVLDD144 pKa = 5.1GITILNAPKK153 pKa = 10.49NSFSVNYY160 pKa = 10.06VEE162 pKa = 6.1NLVLQNIVINNTAGDD177 pKa = 4.08EE178 pKa = 4.19LNSDD182 pKa = 4.34GKK184 pKa = 8.85TLGHH188 pKa = 6.01NTDD191 pKa = 4.18AFDD194 pKa = 4.58INNCDD199 pKa = 2.99GVLFQNITVYY209 pKa = 10.99NQDD212 pKa = 3.01DD213 pKa = 4.16CVAVNSGEE221 pKa = 4.02NVVFRR226 pKa = 11.84DD227 pKa = 3.78ALCSGGHH234 pKa = 6.67GISIGSVGGRR244 pKa = 11.84SNNVVNNITFDD255 pKa = 3.54NVLMQNSQQSVRR267 pKa = 11.84IKK269 pKa = 10.09TIADD273 pKa = 3.51TNGTVSNVTYY283 pKa = 10.49RR284 pKa = 11.84NIVIDD289 pKa = 4.22SPTDD293 pKa = 3.25NTDD296 pKa = 2.85YY297 pKa = 11.44GIIVSQSYY305 pKa = 10.77NGVDD309 pKa = 3.62GEE311 pKa = 4.39PTNGVIISNFVLQNVTGTVYY331 pKa = 10.88EE332 pKa = 4.19DD333 pKa = 4.61AINIYY338 pKa = 9.48IEE340 pKa = 4.62CGEE343 pKa = 4.52GSCIDD348 pKa = 3.69WTWTDD353 pKa = 3.49VNVSGGKK360 pKa = 10.1DD361 pKa = 3.0SADD364 pKa = 3.54CMNIPDD370 pKa = 5.8GISCC374 pKa = 4.99
Molecular weight: 39.5 kDa
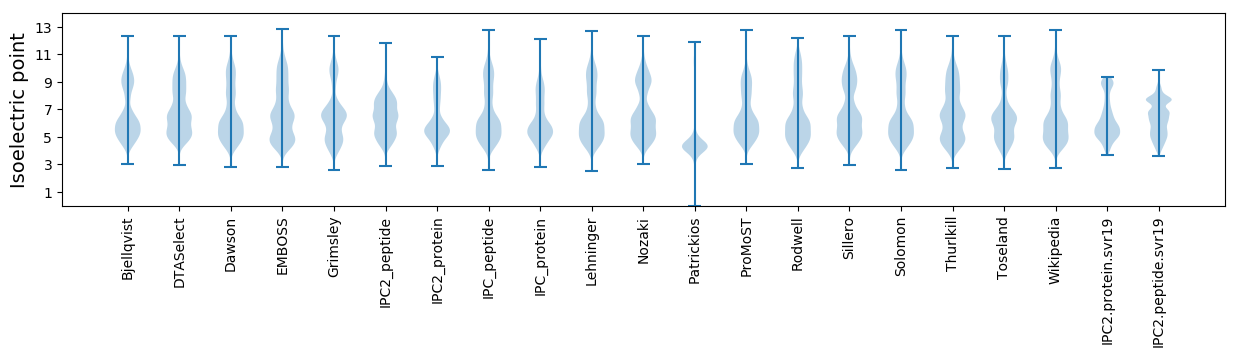
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A423WNU7|A0A423WNU7_9PEZI Uncharacterized protein OS=Valsa sordida OX=252740 GN=VSDG_00025 PE=4 SV=1
MM1 pKa = 7.55PASIHH6 pKa = 6.89CEE8 pKa = 3.56MAQCQPLCPEE18 pKa = 5.44DD19 pKa = 5.42DD20 pKa = 5.01LISTLIAAASHH31 pKa = 7.23RR32 pKa = 11.84YY33 pKa = 7.04TPAAPSPLNPRR44 pKa = 11.84SSIEE48 pKa = 3.86PKK50 pKa = 9.75SRR52 pKa = 11.84KK53 pKa = 8.74HH54 pKa = 5.88APHH57 pKa = 6.41HH58 pKa = 5.49RR59 pKa = 11.84RR60 pKa = 11.84PRR62 pKa = 11.84SSRR65 pKa = 11.84RR66 pKa = 11.84KK67 pKa = 8.26PAEE70 pKa = 3.93TPTEE74 pKa = 3.63RR75 pKa = 11.84LLRR78 pKa = 11.84RR79 pKa = 11.84KK80 pKa = 9.39AALAYY85 pKa = 9.47EE86 pKa = 4.27RR87 pKa = 11.84TTRR90 pKa = 11.84LSPSLSPTHH99 pKa = 5.92STPPPPPPPRR109 pKa = 11.84RR110 pKa = 11.84KK111 pKa = 10.1NGGTVVAARR120 pKa = 11.84PCSSPEE126 pKa = 3.6KK127 pKa = 10.45GPAISCVANEE137 pKa = 4.16ARR139 pKa = 11.84RR140 pKa = 11.84AGQTRR145 pKa = 11.84DD146 pKa = 3.14KK147 pKa = 10.44PLAVEE152 pKa = 3.81WWEE155 pKa = 3.66IRR157 pKa = 11.84HH158 pKa = 6.16RR159 pKa = 11.84LLADD163 pKa = 3.43VEE165 pKa = 4.43GQPAAQLSGVRR176 pKa = 11.84QRR178 pKa = 11.84PASRR182 pKa = 11.84QRR184 pKa = 11.84LEE186 pKa = 3.84ILLMLVLLSACLTLLTVVGVDD207 pKa = 3.52SLKK210 pKa = 10.39GHH212 pKa = 6.76YY213 pKa = 9.41IAGG216 pKa = 3.81
MM1 pKa = 7.55PASIHH6 pKa = 6.89CEE8 pKa = 3.56MAQCQPLCPEE18 pKa = 5.44DD19 pKa = 5.42DD20 pKa = 5.01LISTLIAAASHH31 pKa = 7.23RR32 pKa = 11.84YY33 pKa = 7.04TPAAPSPLNPRR44 pKa = 11.84SSIEE48 pKa = 3.86PKK50 pKa = 9.75SRR52 pKa = 11.84KK53 pKa = 8.74HH54 pKa = 5.88APHH57 pKa = 6.41HH58 pKa = 5.49RR59 pKa = 11.84RR60 pKa = 11.84PRR62 pKa = 11.84SSRR65 pKa = 11.84RR66 pKa = 11.84KK67 pKa = 8.26PAEE70 pKa = 3.93TPTEE74 pKa = 3.63RR75 pKa = 11.84LLRR78 pKa = 11.84RR79 pKa = 11.84KK80 pKa = 9.39AALAYY85 pKa = 9.47EE86 pKa = 4.27RR87 pKa = 11.84TTRR90 pKa = 11.84LSPSLSPTHH99 pKa = 5.92STPPPPPPPRR109 pKa = 11.84RR110 pKa = 11.84KK111 pKa = 10.1NGGTVVAARR120 pKa = 11.84PCSSPEE126 pKa = 3.6KK127 pKa = 10.45GPAISCVANEE137 pKa = 4.16ARR139 pKa = 11.84RR140 pKa = 11.84AGQTRR145 pKa = 11.84DD146 pKa = 3.14KK147 pKa = 10.44PLAVEE152 pKa = 3.81WWEE155 pKa = 3.66IRR157 pKa = 11.84HH158 pKa = 6.16RR159 pKa = 11.84LLADD163 pKa = 3.43VEE165 pKa = 4.43GQPAAQLSGVRR176 pKa = 11.84QRR178 pKa = 11.84PASRR182 pKa = 11.84QRR184 pKa = 11.84LEE186 pKa = 3.84ILLMLVLLSACLTLLTVVGVDD207 pKa = 3.52SLKK210 pKa = 10.39GHH212 pKa = 6.76YY213 pKa = 9.41IAGG216 pKa = 3.81
Molecular weight: 23.65 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4954655 |
50 |
5484 |
504.7 |
55.56 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.151 ± 0.027 | 1.151 ± 0.009 |
5.993 ± 0.018 | 6.249 ± 0.024 |
3.535 ± 0.014 | 7.422 ± 0.025 |
2.334 ± 0.01 | 4.496 ± 0.015 |
4.707 ± 0.024 | 8.596 ± 0.025 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.294 ± 0.009 | 3.503 ± 0.012 |
6.14 ± 0.026 | 3.976 ± 0.017 |
6.126 ± 0.021 | 7.956 ± 0.029 |
5.915 ± 0.016 | 6.282 ± 0.02 |
1.457 ± 0.008 | 2.715 ± 0.014 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
