
Mycoplasma sp. OR1901
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Tenericutes; Mollicutes; Mycoplasmatales; Mycoplasmataceae; Mycoplasma; unclassified Mycoplasma
Average proteome isoelectric point is 6.97
Get precalculated fractions of proteins
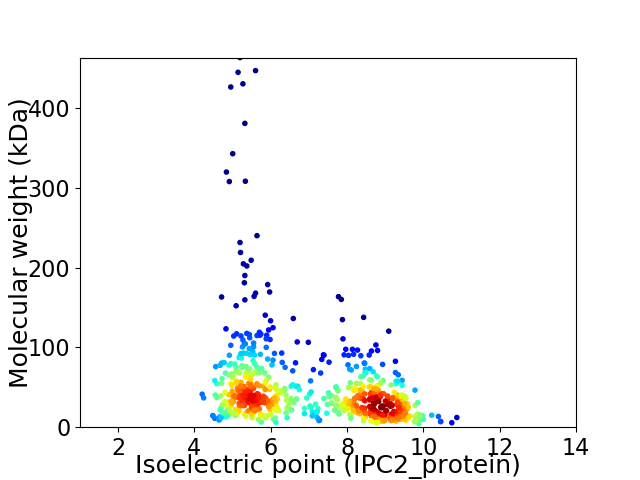
Virtual 2D-PAGE plot for 651 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6N1DVW2|A0A6N1DVW2_9MOLU 50S ribosomal protein L33 OS=Mycoplasma sp. OR1901 OX=2742195 GN=rpmG PE=3 SV=1
MM1 pKa = 7.62KK2 pKa = 10.4LNKK5 pKa = 9.59KK6 pKa = 7.61VLKK9 pKa = 10.59NILITLSGISVVTTVSCSQSQAKK32 pKa = 9.76QEE34 pKa = 4.19VKK36 pKa = 10.73KK37 pKa = 10.61EE38 pKa = 4.19DD39 pKa = 3.63NSGTNADD46 pKa = 3.81TNVVKK51 pKa = 10.83EE52 pKa = 4.2NDD54 pKa = 3.7STSDD58 pKa = 3.28NVNNNSSNEE67 pKa = 3.82EE68 pKa = 4.06STTDD72 pKa = 3.22STNDD76 pKa = 2.94SKK78 pKa = 10.28TKK80 pKa = 10.35VEE82 pKa = 4.18EE83 pKa = 4.43EE84 pKa = 4.42EE85 pKa = 4.28VTTPADD91 pKa = 3.51STNEE95 pKa = 3.76RR96 pKa = 11.84TTEE99 pKa = 4.1GTNSDD104 pKa = 3.25SSSRR108 pKa = 11.84EE109 pKa = 4.05GEE111 pKa = 3.94NTTTDD116 pKa = 3.14EE117 pKa = 4.34KK118 pKa = 10.52QTEE121 pKa = 4.39EE122 pKa = 3.97VDD124 pKa = 4.22SNSSTGEE131 pKa = 3.93TSSDD135 pKa = 3.24GSGSTSGDD143 pKa = 3.11NSSTGEE149 pKa = 3.89ASSSEE154 pKa = 3.88GDD156 pKa = 3.68STSEE160 pKa = 3.99GSKK163 pKa = 10.27SDD165 pKa = 3.34SSEE168 pKa = 4.28GEE170 pKa = 4.19TSSSEE175 pKa = 4.41SEE177 pKa = 4.11STPDD181 pKa = 4.46GEE183 pKa = 4.63KK184 pKa = 9.13QTEE187 pKa = 4.44GTGSSNGEE195 pKa = 4.12TPSSGEE201 pKa = 3.87GTTTGEE207 pKa = 4.23KK208 pKa = 8.39QTGGADD214 pKa = 3.42SDD216 pKa = 4.14SSGEE220 pKa = 4.03VTSPDD225 pKa = 3.35EE226 pKa = 4.65GEE228 pKa = 4.14SGGDD232 pKa = 3.48GEE234 pKa = 4.87TEE236 pKa = 3.93GTIGSNTDD244 pKa = 3.23DD245 pKa = 4.5SGKK248 pKa = 10.41EE249 pKa = 3.8NQKK252 pKa = 9.08QQVKK256 pKa = 9.09VFVVNPKK263 pKa = 10.08LNKK266 pKa = 9.77EE267 pKa = 3.94KK268 pKa = 9.99MIEE271 pKa = 4.18IINSKK276 pKa = 10.07LKK278 pKa = 10.09NGQVKK283 pKa = 10.29LYY285 pKa = 10.24KK286 pKa = 10.11YY287 pKa = 11.08ANTEE291 pKa = 3.63YY292 pKa = 10.46FGVYY296 pKa = 9.69RR297 pKa = 11.84GGSNEE302 pKa = 3.58EE303 pKa = 3.48NRR305 pKa = 11.84FFKK308 pKa = 10.81FDD310 pKa = 3.6VNYY313 pKa = 10.84SEE315 pKa = 5.93SNVKK319 pKa = 10.22LDD321 pKa = 3.54YY322 pKa = 10.93PEE324 pKa = 3.99NAKK327 pKa = 9.3TKK329 pKa = 9.72TFLGNWDD336 pKa = 3.56ADD338 pKa = 3.87KK339 pKa = 11.19KK340 pKa = 11.2VLTISYY346 pKa = 9.26KK347 pKa = 10.36YY348 pKa = 10.2KK349 pKa = 10.74EE350 pKa = 4.1EE351 pKa = 4.2TIEE354 pKa = 4.44QSFNLSNDD362 pKa = 2.86EE363 pKa = 4.18STTDD367 pKa = 3.44EE368 pKa = 4.65NGSEE372 pKa = 4.39DD373 pKa = 3.72SSSSSSSSSSSSEE386 pKa = 3.72VSNNN390 pKa = 2.88
MM1 pKa = 7.62KK2 pKa = 10.4LNKK5 pKa = 9.59KK6 pKa = 7.61VLKK9 pKa = 10.59NILITLSGISVVTTVSCSQSQAKK32 pKa = 9.76QEE34 pKa = 4.19VKK36 pKa = 10.73KK37 pKa = 10.61EE38 pKa = 4.19DD39 pKa = 3.63NSGTNADD46 pKa = 3.81TNVVKK51 pKa = 10.83EE52 pKa = 4.2NDD54 pKa = 3.7STSDD58 pKa = 3.28NVNNNSSNEE67 pKa = 3.82EE68 pKa = 4.06STTDD72 pKa = 3.22STNDD76 pKa = 2.94SKK78 pKa = 10.28TKK80 pKa = 10.35VEE82 pKa = 4.18EE83 pKa = 4.43EE84 pKa = 4.42EE85 pKa = 4.28VTTPADD91 pKa = 3.51STNEE95 pKa = 3.76RR96 pKa = 11.84TTEE99 pKa = 4.1GTNSDD104 pKa = 3.25SSSRR108 pKa = 11.84EE109 pKa = 4.05GEE111 pKa = 3.94NTTTDD116 pKa = 3.14EE117 pKa = 4.34KK118 pKa = 10.52QTEE121 pKa = 4.39EE122 pKa = 3.97VDD124 pKa = 4.22SNSSTGEE131 pKa = 3.93TSSDD135 pKa = 3.24GSGSTSGDD143 pKa = 3.11NSSTGEE149 pKa = 3.89ASSSEE154 pKa = 3.88GDD156 pKa = 3.68STSEE160 pKa = 3.99GSKK163 pKa = 10.27SDD165 pKa = 3.34SSEE168 pKa = 4.28GEE170 pKa = 4.19TSSSEE175 pKa = 4.41SEE177 pKa = 4.11STPDD181 pKa = 4.46GEE183 pKa = 4.63KK184 pKa = 9.13QTEE187 pKa = 4.44GTGSSNGEE195 pKa = 4.12TPSSGEE201 pKa = 3.87GTTTGEE207 pKa = 4.23KK208 pKa = 8.39QTGGADD214 pKa = 3.42SDD216 pKa = 4.14SSGEE220 pKa = 4.03VTSPDD225 pKa = 3.35EE226 pKa = 4.65GEE228 pKa = 4.14SGGDD232 pKa = 3.48GEE234 pKa = 4.87TEE236 pKa = 3.93GTIGSNTDD244 pKa = 3.23DD245 pKa = 4.5SGKK248 pKa = 10.41EE249 pKa = 3.8NQKK252 pKa = 9.08QQVKK256 pKa = 9.09VFVVNPKK263 pKa = 10.08LNKK266 pKa = 9.77EE267 pKa = 3.94KK268 pKa = 9.99MIEE271 pKa = 4.18IINSKK276 pKa = 10.07LKK278 pKa = 10.09NGQVKK283 pKa = 10.29LYY285 pKa = 10.24KK286 pKa = 10.11YY287 pKa = 11.08ANTEE291 pKa = 3.63YY292 pKa = 10.46FGVYY296 pKa = 9.69RR297 pKa = 11.84GGSNEE302 pKa = 3.58EE303 pKa = 3.48NRR305 pKa = 11.84FFKK308 pKa = 10.81FDD310 pKa = 3.6VNYY313 pKa = 10.84SEE315 pKa = 5.93SNVKK319 pKa = 10.22LDD321 pKa = 3.54YY322 pKa = 10.93PEE324 pKa = 3.99NAKK327 pKa = 9.3TKK329 pKa = 9.72TFLGNWDD336 pKa = 3.56ADD338 pKa = 3.87KK339 pKa = 11.19KK340 pKa = 11.2VLTISYY346 pKa = 9.26KK347 pKa = 10.36YY348 pKa = 10.2KK349 pKa = 10.74EE350 pKa = 4.1EE351 pKa = 4.2TIEE354 pKa = 4.44QSFNLSNDD362 pKa = 2.86EE363 pKa = 4.18STTDD367 pKa = 3.44EE368 pKa = 4.65NGSEE372 pKa = 4.39DD373 pKa = 3.72SSSSSSSSSSSSEE386 pKa = 3.72VSNNN390 pKa = 2.88
Molecular weight: 41.37 kDa
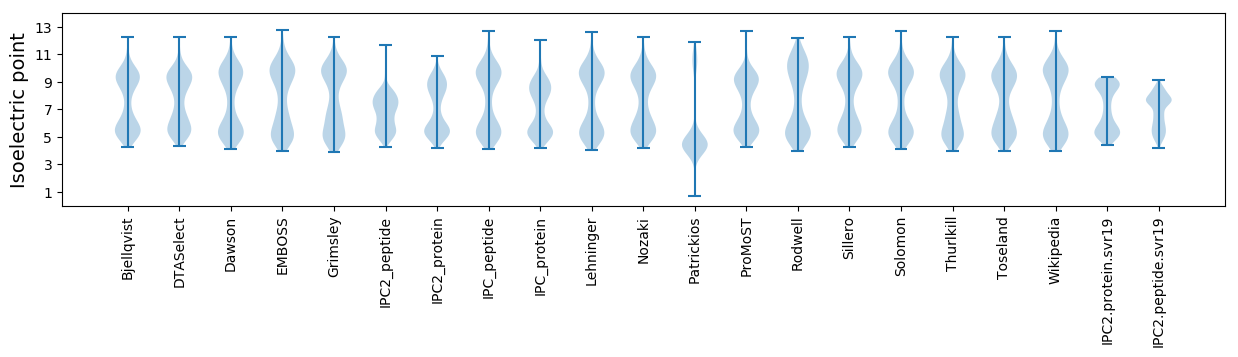
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6N1DSX4|A0A6N1DSX4_9MOLU 2 3-bisphosphoglycerate-independent phosphoglycerate mutase OS=Mycoplasma sp. OR1901 OX=2742195 GN=gpmI PE=3 SV=1
MM1 pKa = 7.94AYY3 pKa = 10.34NKK5 pKa = 9.78RR6 pKa = 11.84KK7 pKa = 9.92KK8 pKa = 10.1VFSGRR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84VCSFCEE21 pKa = 4.22SKK23 pKa = 10.54SQYY26 pKa = 10.55VDD28 pKa = 3.44YY29 pKa = 11.33KK30 pKa = 11.13DD31 pKa = 5.0VEE33 pKa = 4.36LLNNFISATGQIKK46 pKa = 10.25ARR48 pKa = 11.84SITGTCAKK56 pKa = 9.88HH57 pKa = 4.71QRR59 pKa = 11.84KK60 pKa = 8.97VSNAIKK66 pKa = 9.97RR67 pKa = 11.84ARR69 pKa = 11.84FIAFMPYY76 pKa = 8.59TVVRR80 pKa = 11.84VRR82 pKa = 11.84NLSKK86 pKa = 11.05
MM1 pKa = 7.94AYY3 pKa = 10.34NKK5 pKa = 9.78RR6 pKa = 11.84KK7 pKa = 9.92KK8 pKa = 10.1VFSGRR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84VCSFCEE21 pKa = 4.22SKK23 pKa = 10.54SQYY26 pKa = 10.55VDD28 pKa = 3.44YY29 pKa = 11.33KK30 pKa = 11.13DD31 pKa = 5.0VEE33 pKa = 4.36LLNNFISATGQIKK46 pKa = 10.25ARR48 pKa = 11.84SITGTCAKK56 pKa = 9.88HH57 pKa = 4.71QRR59 pKa = 11.84KK60 pKa = 8.97VSNAIKK66 pKa = 9.97RR67 pKa = 11.84ARR69 pKa = 11.84FIAFMPYY76 pKa = 8.59TVVRR80 pKa = 11.84VRR82 pKa = 11.84NLSKK86 pKa = 11.05
Molecular weight: 9.97 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
288149 |
37 |
4018 |
442.6 |
50.82 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.728 ± 0.13 | 0.254 ± 0.017 |
6.074 ± 0.075 | 7.67 ± 0.145 |
5.011 ± 0.122 | 4.455 ± 0.123 |
1.29 ± 0.035 | 9.032 ± 0.105 |
10.534 ± 0.103 | 9.178 ± 0.085 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.542 ± 0.051 | 8.968 ± 0.146 |
2.435 ± 0.069 | 3.22 ± 0.078 |
2.892 ± 0.054 | 6.771 ± 0.081 |
5.453 ± 0.063 | 5.516 ± 0.073 |
0.845 ± 0.029 | 4.132 ± 0.1 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
