
Albimonas pacifica
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Rhodobacteraceae; Albimonas
Average proteome isoelectric point is 6.27
Get precalculated fractions of proteins
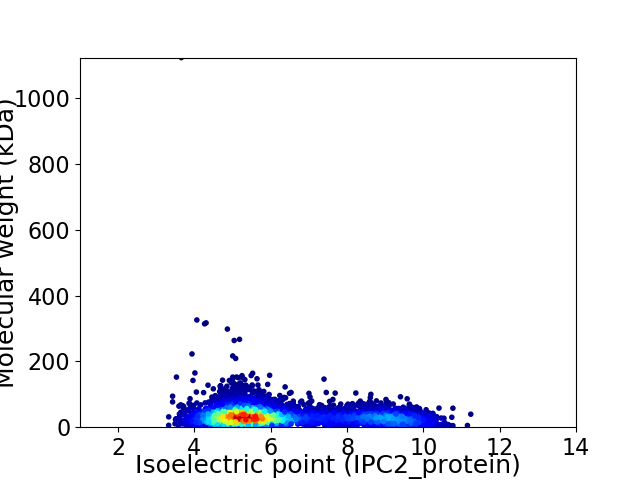
Virtual 2D-PAGE plot for 5319 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1I3F5L2|A0A1I3F5L2_9RHOB Two-component system OmpR family phosphate regulon response regulator OmpR OS=Albimonas pacifica OX=1114924 GN=SAMN05216258_10478 PE=4 SV=1
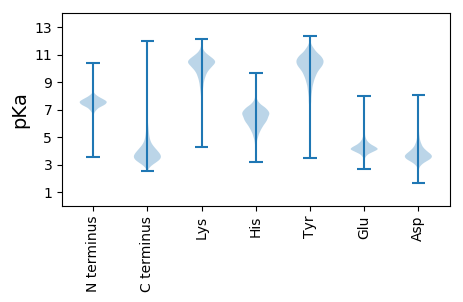
MM1 pKa = 7.54TYY3 pKa = 10.78GYY5 pKa = 10.56LNYY8 pKa = 10.55GSGSGSFSGYY18 pKa = 10.41SKK20 pKa = 10.61GHH22 pKa = 6.35GGSGHH27 pKa = 6.62DD28 pKa = 3.24AWGSKK33 pKa = 9.66GGYY36 pKa = 8.51GGSSGGHH43 pKa = 6.1GGSSGGWGSSGGSSGGWGGSKK64 pKa = 10.38DD65 pKa = 3.43HH66 pKa = 6.8GGSKK70 pKa = 10.56GGSSWGSSSSWGDD83 pKa = 3.32KK84 pKa = 10.55DD85 pKa = 3.61DD86 pKa = 4.97CGWGGSKK93 pKa = 10.5GGSGWGGSKK102 pKa = 10.53GGSSWEE108 pKa = 3.98SCGAKK113 pKa = 9.86GGYY116 pKa = 9.35GGSKK120 pKa = 10.39GGASWGGKK128 pKa = 9.02DD129 pKa = 3.26DD130 pKa = 4.76HH131 pKa = 6.75GWGGKK136 pKa = 9.24GGCGPDD142 pKa = 3.29EE143 pKa = 4.31TPEE146 pKa = 4.03FEE148 pKa = 4.99PGAEE152 pKa = 4.04VAQCSFTLEE161 pKa = 3.86LDD163 pKa = 3.56GVKK166 pKa = 10.41VVVTVTQLEE175 pKa = 4.04NDD177 pKa = 3.59ALRR180 pKa = 11.84FDD182 pKa = 4.38VVVADD187 pKa = 4.04DD188 pKa = 3.66AARR191 pKa = 11.84IGDD194 pKa = 3.71LRR196 pKa = 11.84GLFFNVDD203 pKa = 3.74DD204 pKa = 4.64GVEE207 pKa = 4.05GDD209 pKa = 3.68LAIYY213 pKa = 10.33GDD215 pKa = 5.06DD216 pKa = 3.5VTEE219 pKa = 4.26SKK221 pKa = 10.65FGHH224 pKa = 6.3EE225 pKa = 4.57SVNDD229 pKa = 3.79LGNGVNVKK237 pKa = 8.74GTGAAYY243 pKa = 10.15DD244 pKa = 3.53VGVEE248 pKa = 4.46FGSSGVGADD257 pKa = 4.04DD258 pKa = 3.68VRR260 pKa = 11.84STTFYY265 pKa = 11.21LVNLDD270 pKa = 3.9GALTLDD276 pKa = 4.26DD277 pKa = 5.39LADD280 pKa = 3.43QGVAARR286 pKa = 11.84LTSVGEE292 pKa = 3.82EE293 pKa = 3.95GGRR296 pKa = 11.84RR297 pKa = 11.84EE298 pKa = 4.69DD299 pKa = 3.37SLKK302 pKa = 9.46ITGEE306 pKa = 4.09TGALDD311 pKa = 3.54CGGFNNAAPPEE322 pKa = 4.37PADD325 pKa = 4.39DD326 pKa = 4.05AAAVCAGDD334 pKa = 3.92QVTIDD339 pKa = 4.47LLANDD344 pKa = 5.41LDD346 pKa = 4.35PGEE349 pKa = 4.29TDD351 pKa = 5.48FGAEE355 pKa = 4.28EE356 pKa = 4.0NWGLTIAAVFTATGAQYY373 pKa = 10.42FADD376 pKa = 4.77LAPGEE381 pKa = 4.67WITLDD386 pKa = 2.89SGARR390 pKa = 11.84VTLVDD395 pKa = 3.84GEE397 pKa = 4.62LVYY400 pKa = 11.14DD401 pKa = 4.41SAGVWDD407 pKa = 4.76DD408 pKa = 3.89LLIGEE413 pKa = 4.8HH414 pKa = 6.71AVDD417 pKa = 3.44TFEE420 pKa = 4.47YY421 pKa = 10.9AVFDD425 pKa = 3.73SDD427 pKa = 3.94GAPGFAEE434 pKa = 3.99VSVTIGGALNLVEE447 pKa = 5.17TIGNDD452 pKa = 3.45DD453 pKa = 3.89PASAFVDD460 pKa = 3.97LLGLPGGGGITGAFDD475 pKa = 3.85FAGDD479 pKa = 3.75LAGLGGGYY487 pKa = 6.91TQIYY491 pKa = 9.71CIDD494 pKa = 3.68RR495 pKa = 11.84DD496 pKa = 3.88RR497 pKa = 11.84PLNTAPTDD505 pKa = 3.55VLVYY509 pKa = 10.68SSLDD513 pKa = 3.37ADD515 pKa = 4.18GLPTTSFDD523 pKa = 3.35GGSFVDD529 pKa = 3.87NPEE532 pKa = 4.03NLDD535 pKa = 3.63LVNWILNQGYY545 pKa = 10.09EE546 pKa = 4.21GTYY549 pKa = 10.03SYY551 pKa = 11.9NDD553 pKa = 3.23IQSAIWQLVDD563 pKa = 3.69DD564 pKa = 4.97RR565 pKa = 11.84GGIDD569 pKa = 3.32TLIFTPGIQLSAGAAAIVAAAQAEE593 pKa = 4.75GEE595 pKa = 4.57GFVPDD600 pKa = 4.21ADD602 pKa = 4.04LGQTVGMIFQPVSGSEE618 pKa = 3.81EE619 pKa = 3.65SGYY622 pKa = 10.93VSNGQIFIAGFQLEE636 pKa = 4.34DD637 pKa = 4.09CDD639 pKa = 4.91AHH641 pKa = 7.35MLMM644 pKa = 5.47
MM1 pKa = 7.54TYY3 pKa = 10.78GYY5 pKa = 10.56LNYY8 pKa = 10.55GSGSGSFSGYY18 pKa = 10.41SKK20 pKa = 10.61GHH22 pKa = 6.35GGSGHH27 pKa = 6.62DD28 pKa = 3.24AWGSKK33 pKa = 9.66GGYY36 pKa = 8.51GGSSGGHH43 pKa = 6.1GGSSGGWGSSGGSSGGWGGSKK64 pKa = 10.38DD65 pKa = 3.43HH66 pKa = 6.8GGSKK70 pKa = 10.56GGSSWGSSSSWGDD83 pKa = 3.32KK84 pKa = 10.55DD85 pKa = 3.61DD86 pKa = 4.97CGWGGSKK93 pKa = 10.5GGSGWGGSKK102 pKa = 10.53GGSSWEE108 pKa = 3.98SCGAKK113 pKa = 9.86GGYY116 pKa = 9.35GGSKK120 pKa = 10.39GGASWGGKK128 pKa = 9.02DD129 pKa = 3.26DD130 pKa = 4.76HH131 pKa = 6.75GWGGKK136 pKa = 9.24GGCGPDD142 pKa = 3.29EE143 pKa = 4.31TPEE146 pKa = 4.03FEE148 pKa = 4.99PGAEE152 pKa = 4.04VAQCSFTLEE161 pKa = 3.86LDD163 pKa = 3.56GVKK166 pKa = 10.41VVVTVTQLEE175 pKa = 4.04NDD177 pKa = 3.59ALRR180 pKa = 11.84FDD182 pKa = 4.38VVVADD187 pKa = 4.04DD188 pKa = 3.66AARR191 pKa = 11.84IGDD194 pKa = 3.71LRR196 pKa = 11.84GLFFNVDD203 pKa = 3.74DD204 pKa = 4.64GVEE207 pKa = 4.05GDD209 pKa = 3.68LAIYY213 pKa = 10.33GDD215 pKa = 5.06DD216 pKa = 3.5VTEE219 pKa = 4.26SKK221 pKa = 10.65FGHH224 pKa = 6.3EE225 pKa = 4.57SVNDD229 pKa = 3.79LGNGVNVKK237 pKa = 8.74GTGAAYY243 pKa = 10.15DD244 pKa = 3.53VGVEE248 pKa = 4.46FGSSGVGADD257 pKa = 4.04DD258 pKa = 3.68VRR260 pKa = 11.84STTFYY265 pKa = 11.21LVNLDD270 pKa = 3.9GALTLDD276 pKa = 4.26DD277 pKa = 5.39LADD280 pKa = 3.43QGVAARR286 pKa = 11.84LTSVGEE292 pKa = 3.82EE293 pKa = 3.95GGRR296 pKa = 11.84RR297 pKa = 11.84EE298 pKa = 4.69DD299 pKa = 3.37SLKK302 pKa = 9.46ITGEE306 pKa = 4.09TGALDD311 pKa = 3.54CGGFNNAAPPEE322 pKa = 4.37PADD325 pKa = 4.39DD326 pKa = 4.05AAAVCAGDD334 pKa = 3.92QVTIDD339 pKa = 4.47LLANDD344 pKa = 5.41LDD346 pKa = 4.35PGEE349 pKa = 4.29TDD351 pKa = 5.48FGAEE355 pKa = 4.28EE356 pKa = 4.0NWGLTIAAVFTATGAQYY373 pKa = 10.42FADD376 pKa = 4.77LAPGEE381 pKa = 4.67WITLDD386 pKa = 2.89SGARR390 pKa = 11.84VTLVDD395 pKa = 3.84GEE397 pKa = 4.62LVYY400 pKa = 11.14DD401 pKa = 4.41SAGVWDD407 pKa = 4.76DD408 pKa = 3.89LLIGEE413 pKa = 4.8HH414 pKa = 6.71AVDD417 pKa = 3.44TFEE420 pKa = 4.47YY421 pKa = 10.9AVFDD425 pKa = 3.73SDD427 pKa = 3.94GAPGFAEE434 pKa = 3.99VSVTIGGALNLVEE447 pKa = 5.17TIGNDD452 pKa = 3.45DD453 pKa = 3.89PASAFVDD460 pKa = 3.97LLGLPGGGGITGAFDD475 pKa = 3.85FAGDD479 pKa = 3.75LAGLGGGYY487 pKa = 6.91TQIYY491 pKa = 9.71CIDD494 pKa = 3.68RR495 pKa = 11.84DD496 pKa = 3.88RR497 pKa = 11.84PLNTAPTDD505 pKa = 3.55VLVYY509 pKa = 10.68SSLDD513 pKa = 3.37ADD515 pKa = 4.18GLPTTSFDD523 pKa = 3.35GGSFVDD529 pKa = 3.87NPEE532 pKa = 4.03NLDD535 pKa = 3.63LVNWILNQGYY545 pKa = 10.09EE546 pKa = 4.21GTYY549 pKa = 10.03SYY551 pKa = 11.9NDD553 pKa = 3.23IQSAIWQLVDD563 pKa = 3.69DD564 pKa = 4.97RR565 pKa = 11.84GGIDD569 pKa = 3.32TLIFTPGIQLSAGAAAIVAAAQAEE593 pKa = 4.75GEE595 pKa = 4.57GFVPDD600 pKa = 4.21ADD602 pKa = 4.04LGQTVGMIFQPVSGSEE618 pKa = 3.81EE619 pKa = 3.65SGYY622 pKa = 10.93VSNGQIFIAGFQLEE636 pKa = 4.34DD637 pKa = 4.09CDD639 pKa = 4.91AHH641 pKa = 7.35MLMM644 pKa = 5.47
Molecular weight: 65.28 kDa
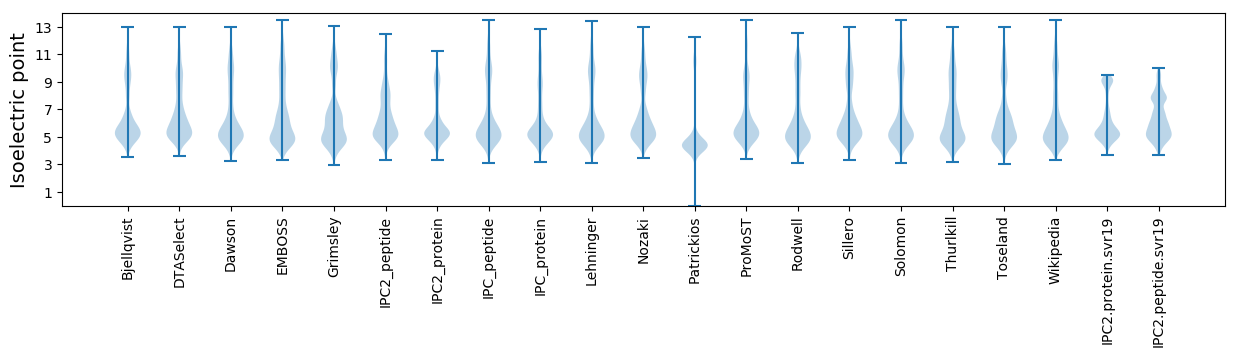
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1I3IHR2|A0A1I3IHR2_9RHOB Basal-body rod modification protein FlgD OS=Albimonas pacifica OX=1114924 GN=SAMN05216258_10761 PE=3 SV=1
MM1 pKa = 7.71RR2 pKa = 11.84PCPISRR8 pKa = 11.84SRR10 pKa = 11.84TPRR13 pKa = 11.84PVPSPPRR20 pKa = 11.84CGGARR25 pKa = 11.84RR26 pKa = 11.84RR27 pKa = 11.84LRR29 pKa = 11.84ARR31 pKa = 11.84PRR33 pKa = 11.84AQRR36 pKa = 11.84RR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84SRR41 pKa = 11.84TPAVPIGRR49 pKa = 11.84GGGPRR54 pKa = 11.84RR55 pKa = 11.84WSRR58 pKa = 11.84PPNWTLRR65 pKa = 11.84PRR67 pKa = 11.84RR68 pKa = 11.84WRR70 pKa = 11.84STWRR74 pKa = 11.84SRR76 pKa = 11.84PPRR79 pKa = 11.84RR80 pKa = 11.84RR81 pKa = 11.84RR82 pKa = 11.84NRR84 pKa = 11.84RR85 pKa = 11.84PNPRR89 pKa = 11.84HH90 pKa = 5.82PLPGRR95 pKa = 11.84RR96 pKa = 11.84LRR98 pKa = 11.84AVPSPRR104 pKa = 11.84RR105 pKa = 11.84RR106 pKa = 11.84GRR108 pKa = 11.84PRR110 pKa = 11.84RR111 pKa = 11.84RR112 pKa = 11.84PRR114 pKa = 11.84PRR116 pKa = 11.84SSVGHH121 pKa = 5.1PLKK124 pKa = 10.65RR125 pKa = 11.84RR126 pKa = 11.84GRR128 pKa = 11.84GPPPPRR134 pKa = 11.84LVAPPWTAVQARR146 pKa = 11.84PFRR149 pKa = 11.84KK150 pKa = 8.44ATPSCRR156 pKa = 11.84PLARR160 pKa = 11.84RR161 pKa = 11.84GQTTLRR167 pKa = 11.84RR168 pKa = 11.84RR169 pKa = 11.84RR170 pKa = 11.84FRR172 pKa = 11.84SCPPRR177 pKa = 11.84PPLQTGGRR185 pKa = 11.84KK186 pKa = 9.15GKK188 pKa = 9.34RR189 pKa = 11.84RR190 pKa = 11.84RR191 pKa = 11.84WRR193 pKa = 11.84MPRR196 pKa = 11.84AVHH199 pKa = 6.71RR200 pKa = 11.84SAARR204 pKa = 11.84TLRR207 pKa = 11.84HH208 pKa = 4.81QAPRR212 pKa = 11.84RR213 pKa = 11.84ASRR216 pKa = 11.84PTRR219 pKa = 11.84ALLPCRR225 pKa = 11.84RR226 pKa = 11.84RR227 pKa = 11.84RR228 pKa = 11.84PTRR231 pKa = 11.84GSWNGRR237 pKa = 11.84PRR239 pKa = 11.84WLLRR243 pKa = 11.84PWRR246 pKa = 11.84RR247 pKa = 11.84HH248 pKa = 4.38GRR250 pKa = 11.84PTAAPDD256 pKa = 3.93RR257 pKa = 11.84PGSWTRR263 pKa = 11.84SARR266 pKa = 11.84RR267 pKa = 11.84ARR269 pKa = 11.84RR270 pKa = 11.84RR271 pKa = 11.84RR272 pKa = 11.84PRR274 pKa = 11.84RR275 pKa = 11.84PCNSARR281 pKa = 11.84SPPRR285 pKa = 11.84SGLRR289 pKa = 11.84DD290 pKa = 3.66FRR292 pKa = 11.84RR293 pKa = 11.84RR294 pKa = 11.84PQGRR298 pKa = 11.84RR299 pKa = 11.84RR300 pKa = 11.84RR301 pKa = 11.84AGRR304 pKa = 11.84PFATGGPEE312 pKa = 3.18RR313 pKa = 11.84RR314 pKa = 11.84ARR316 pKa = 11.84SGGKK320 pKa = 9.28RR321 pKa = 11.84PAFARR326 pKa = 11.84RR327 pKa = 11.84RR328 pKa = 11.84ALRR331 pKa = 11.84RR332 pKa = 11.84RR333 pKa = 11.84AA334 pKa = 3.22
MM1 pKa = 7.71RR2 pKa = 11.84PCPISRR8 pKa = 11.84SRR10 pKa = 11.84TPRR13 pKa = 11.84PVPSPPRR20 pKa = 11.84CGGARR25 pKa = 11.84RR26 pKa = 11.84RR27 pKa = 11.84LRR29 pKa = 11.84ARR31 pKa = 11.84PRR33 pKa = 11.84AQRR36 pKa = 11.84RR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84SRR41 pKa = 11.84TPAVPIGRR49 pKa = 11.84GGGPRR54 pKa = 11.84RR55 pKa = 11.84WSRR58 pKa = 11.84PPNWTLRR65 pKa = 11.84PRR67 pKa = 11.84RR68 pKa = 11.84WRR70 pKa = 11.84STWRR74 pKa = 11.84SRR76 pKa = 11.84PPRR79 pKa = 11.84RR80 pKa = 11.84RR81 pKa = 11.84RR82 pKa = 11.84NRR84 pKa = 11.84RR85 pKa = 11.84PNPRR89 pKa = 11.84HH90 pKa = 5.82PLPGRR95 pKa = 11.84RR96 pKa = 11.84LRR98 pKa = 11.84AVPSPRR104 pKa = 11.84RR105 pKa = 11.84RR106 pKa = 11.84GRR108 pKa = 11.84PRR110 pKa = 11.84RR111 pKa = 11.84RR112 pKa = 11.84PRR114 pKa = 11.84PRR116 pKa = 11.84SSVGHH121 pKa = 5.1PLKK124 pKa = 10.65RR125 pKa = 11.84RR126 pKa = 11.84GRR128 pKa = 11.84GPPPPRR134 pKa = 11.84LVAPPWTAVQARR146 pKa = 11.84PFRR149 pKa = 11.84KK150 pKa = 8.44ATPSCRR156 pKa = 11.84PLARR160 pKa = 11.84RR161 pKa = 11.84GQTTLRR167 pKa = 11.84RR168 pKa = 11.84RR169 pKa = 11.84RR170 pKa = 11.84FRR172 pKa = 11.84SCPPRR177 pKa = 11.84PPLQTGGRR185 pKa = 11.84KK186 pKa = 9.15GKK188 pKa = 9.34RR189 pKa = 11.84RR190 pKa = 11.84RR191 pKa = 11.84WRR193 pKa = 11.84MPRR196 pKa = 11.84AVHH199 pKa = 6.71RR200 pKa = 11.84SAARR204 pKa = 11.84TLRR207 pKa = 11.84HH208 pKa = 4.81QAPRR212 pKa = 11.84RR213 pKa = 11.84ASRR216 pKa = 11.84PTRR219 pKa = 11.84ALLPCRR225 pKa = 11.84RR226 pKa = 11.84RR227 pKa = 11.84RR228 pKa = 11.84PTRR231 pKa = 11.84GSWNGRR237 pKa = 11.84PRR239 pKa = 11.84WLLRR243 pKa = 11.84PWRR246 pKa = 11.84RR247 pKa = 11.84HH248 pKa = 4.38GRR250 pKa = 11.84PTAAPDD256 pKa = 3.93RR257 pKa = 11.84PGSWTRR263 pKa = 11.84SARR266 pKa = 11.84RR267 pKa = 11.84ARR269 pKa = 11.84RR270 pKa = 11.84RR271 pKa = 11.84RR272 pKa = 11.84PRR274 pKa = 11.84RR275 pKa = 11.84PCNSARR281 pKa = 11.84SPPRR285 pKa = 11.84SGLRR289 pKa = 11.84DD290 pKa = 3.66FRR292 pKa = 11.84RR293 pKa = 11.84RR294 pKa = 11.84PQGRR298 pKa = 11.84RR299 pKa = 11.84RR300 pKa = 11.84RR301 pKa = 11.84AGRR304 pKa = 11.84PFATGGPEE312 pKa = 3.18RR313 pKa = 11.84RR314 pKa = 11.84ARR316 pKa = 11.84SGGKK320 pKa = 9.28RR321 pKa = 11.84PAFARR326 pKa = 11.84RR327 pKa = 11.84RR328 pKa = 11.84ALRR331 pKa = 11.84RR332 pKa = 11.84RR333 pKa = 11.84AA334 pKa = 3.22
Molecular weight: 39.57 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1748452 |
24 |
11104 |
328.7 |
35.15 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
15.359 ± 0.069 | 0.836 ± 0.012 |
5.726 ± 0.044 | 6.276 ± 0.034 |
3.384 ± 0.019 | 9.482 ± 0.056 |
1.829 ± 0.016 | 4.097 ± 0.029 |
2.243 ± 0.026 | 10.242 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.45 ± 0.021 | 1.805 ± 0.019 |
6.015 ± 0.044 | 2.559 ± 0.019 |
7.979 ± 0.055 | 4.594 ± 0.03 |
4.678 ± 0.03 | 7.128 ± 0.035 |
1.496 ± 0.015 | 1.821 ± 0.014 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
