
Sulfurovum sp. (strain NBC37-1)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; delta/epsilon subdivisions; Epsilonproteobacteria; Campylobacterales; Sulfurovaceae; Sulfurovum; unclassified Sulfurovum
Average proteome isoelectric point is 6.59
Get precalculated fractions of proteins
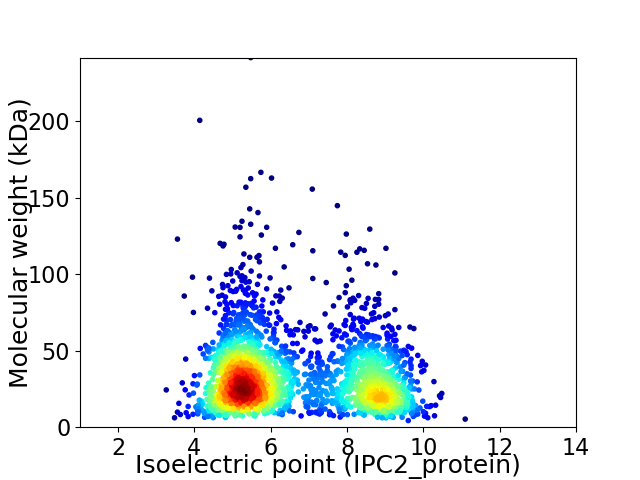
Virtual 2D-PAGE plot for 2433 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|A6QC28|RUVB_SULNB Holliday junction ATP-dependent DNA helicase RuvB OS=Sulfurovum sp. (strain NBC37-1) OX=387093 GN=ruvB PE=3 SV=1
MM1 pKa = 7.57KK2 pKa = 9.17KK3 pKa = 8.57TYY5 pKa = 10.33KK6 pKa = 10.33IILKK10 pKa = 9.93QFLFLGMMLAGSSVSAATLDD30 pKa = 3.6WNTVGWSPAGSLTQSYY46 pKa = 10.13TDD48 pKa = 3.32VDD50 pKa = 4.04SSKK53 pKa = 11.24VNIDD57 pKa = 3.1VTMTEE62 pKa = 3.81DD63 pKa = 3.19TDD65 pKa = 4.06RR66 pKa = 11.84YY67 pKa = 9.63NSGVPSVNSTYY78 pKa = 11.29GLDD81 pKa = 3.53YY82 pKa = 10.73YY83 pKa = 11.72VNYY86 pKa = 10.62SSNTQKK92 pKa = 10.54IKK94 pKa = 8.3TTITFSSPVKK104 pKa = 10.2ISFLRR109 pKa = 11.84WIDD112 pKa = 3.17IDD114 pKa = 3.85SGNPSTDD121 pKa = 3.33FDD123 pKa = 4.07DD124 pKa = 4.83RR125 pKa = 11.84VIVTAKK131 pKa = 8.82DD132 pKa = 3.28TSGATVYY139 pKa = 10.4PSSEE143 pKa = 4.13TLGDD147 pKa = 4.54HH148 pKa = 6.76IDD150 pKa = 3.5KK151 pKa = 10.9NGPGDD156 pKa = 3.89YY157 pKa = 10.53EE158 pKa = 5.12SDD160 pKa = 3.27GTTLNDD166 pKa = 4.83DD167 pKa = 4.6DD168 pKa = 5.13PDD170 pKa = 5.62GYY172 pKa = 10.06VTVAFEE178 pKa = 4.16DD179 pKa = 4.62VYY181 pKa = 10.0VTEE184 pKa = 4.5VSFDD188 pKa = 3.58YY189 pKa = 11.28TNGSNAQSDD198 pKa = 4.21PNNQGIYY205 pKa = 9.64LANITFDD212 pKa = 4.6ALDD215 pKa = 3.7SDD217 pKa = 4.47GDD219 pKa = 4.18GIADD223 pKa = 4.09VKK225 pKa = 11.31DD226 pKa = 3.38IDD228 pKa = 5.15DD229 pKa = 5.13DD230 pKa = 4.31NDD232 pKa = 4.58GILDD236 pKa = 3.74VDD238 pKa = 3.93EE239 pKa = 4.55STPAVTEE246 pKa = 4.35SNTASGTIPDD256 pKa = 3.55NGYY259 pKa = 9.82PNTCLDD265 pKa = 3.69RR266 pKa = 11.84TFDD269 pKa = 3.61IQDD272 pKa = 3.3SGIVNDD278 pKa = 3.51VTIKK282 pKa = 10.72VDD284 pKa = 3.86IAHH287 pKa = 6.84TWRR290 pKa = 11.84NDD292 pKa = 3.83LIVQLISPFGTAVDD306 pKa = 5.06LIRR309 pKa = 11.84NEE311 pKa = 4.33GGSHH315 pKa = 6.3NNLSATFFDD324 pKa = 4.23AATTSIVGDD333 pKa = 3.7TTDD336 pKa = 3.59FTLGSFLPRR345 pKa = 11.84QPEE348 pKa = 3.91QALSAFNGEE357 pKa = 4.55DD358 pKa = 3.66PQGTWTLHH366 pKa = 5.13MCDD369 pKa = 5.31DD370 pKa = 4.65ASQDD374 pKa = 2.98TGTFNEE380 pKa = 4.2ANLTINYY387 pKa = 9.36VNEE390 pKa = 4.33LDD392 pKa = 4.13SDD394 pKa = 3.75NDD396 pKa = 4.77GIIDD400 pKa = 4.69CLDD403 pKa = 4.03LDD405 pKa = 4.48SDD407 pKa = 4.52NDD409 pKa = 4.72GIPDD413 pKa = 3.58NVEE416 pKa = 3.93AQPTATYY423 pKa = 10.48DD424 pKa = 3.7EE425 pKa = 5.6PDD427 pKa = 4.04SAWADD432 pKa = 3.26ADD434 pKa = 3.89GDD436 pKa = 4.24GLADD440 pKa = 4.82QYY442 pKa = 11.38DD443 pKa = 4.08TDD445 pKa = 3.85SGGTAVTPSDD455 pKa = 3.74TDD457 pKa = 3.44GDD459 pKa = 4.59GIPDD463 pKa = 3.91YY464 pKa = 11.22LDD466 pKa = 3.81SDD468 pKa = 4.39SDD470 pKa = 3.74NDD472 pKa = 4.29GYY474 pKa = 9.68TDD476 pKa = 3.84CEE478 pKa = 4.31EE479 pKa = 4.53GNSVATGCPITPGDD493 pKa = 3.67TASTKK498 pKa = 10.13TGLVSWAQGSLGDD511 pKa = 4.52VYY513 pKa = 11.04WDD515 pKa = 3.63TTNSASVSNGNVDD528 pKa = 4.8DD529 pKa = 4.84PSNDD533 pKa = 3.31LFNEE537 pKa = 4.22TGDD540 pKa = 3.68TSEE543 pKa = 3.64VGYY546 pKa = 10.99RR547 pKa = 11.84EE548 pKa = 3.96FLCGKK553 pKa = 9.57NRR555 pKa = 11.84TTLTHH560 pKa = 5.86FQWKK564 pKa = 9.23IVSFCCATGSNHH576 pKa = 6.93IEE578 pKa = 4.19DD579 pKa = 5.0LLGGDD584 pKa = 4.21LGAYY588 pKa = 6.49GTDD591 pKa = 2.21WVVFKK596 pKa = 11.02QSGTDD601 pKa = 3.17QYY603 pKa = 11.16EE604 pKa = 4.15INSGHH609 pKa = 6.84KK610 pKa = 9.0NTTKK614 pKa = 10.73AQLAATDD621 pKa = 3.76TVVPGKK627 pKa = 10.3GYY629 pKa = 9.86WIIADD634 pKa = 3.94LGGAGNEE641 pKa = 4.09KK642 pKa = 10.71NITIDD647 pKa = 3.22KK648 pKa = 9.06TLSNLSPASTVTSSSVGITNPDD670 pKa = 3.01FTEE673 pKa = 3.81VHH675 pKa = 6.68EE676 pKa = 4.27YY677 pKa = 10.89LLPKK681 pKa = 10.7NEE683 pKa = 3.92VSDD686 pKa = 4.66PNTVDD691 pKa = 3.16YY692 pKa = 10.68KK693 pKa = 11.22KK694 pKa = 11.27YY695 pKa = 9.01MAGNPFPYY703 pKa = 10.29AFQLSDD709 pKa = 4.87LYY711 pKa = 10.94FKK713 pKa = 11.07HH714 pKa = 6.28NAGGMSYY721 pKa = 11.3NEE723 pKa = 4.53MGNTANDD730 pKa = 3.19NYY732 pKa = 10.27INKK735 pKa = 9.18IVYY738 pKa = 9.71KK739 pKa = 10.25HH740 pKa = 6.83DD741 pKa = 4.18SNKK744 pKa = 9.25TGPVSGYY751 pKa = 9.09MAVDD755 pKa = 3.8PATPGFDD762 pKa = 4.17GSIQPMEE769 pKa = 4.04GFFIKK774 pKa = 9.94IEE776 pKa = 4.03KK777 pKa = 9.66NQTDD781 pKa = 3.38NYY783 pKa = 9.06VNHH786 pKa = 6.65FAYY789 pKa = 10.0PLMNKK794 pKa = 9.64
MM1 pKa = 7.57KK2 pKa = 9.17KK3 pKa = 8.57TYY5 pKa = 10.33KK6 pKa = 10.33IILKK10 pKa = 9.93QFLFLGMMLAGSSVSAATLDD30 pKa = 3.6WNTVGWSPAGSLTQSYY46 pKa = 10.13TDD48 pKa = 3.32VDD50 pKa = 4.04SSKK53 pKa = 11.24VNIDD57 pKa = 3.1VTMTEE62 pKa = 3.81DD63 pKa = 3.19TDD65 pKa = 4.06RR66 pKa = 11.84YY67 pKa = 9.63NSGVPSVNSTYY78 pKa = 11.29GLDD81 pKa = 3.53YY82 pKa = 10.73YY83 pKa = 11.72VNYY86 pKa = 10.62SSNTQKK92 pKa = 10.54IKK94 pKa = 8.3TTITFSSPVKK104 pKa = 10.2ISFLRR109 pKa = 11.84WIDD112 pKa = 3.17IDD114 pKa = 3.85SGNPSTDD121 pKa = 3.33FDD123 pKa = 4.07DD124 pKa = 4.83RR125 pKa = 11.84VIVTAKK131 pKa = 8.82DD132 pKa = 3.28TSGATVYY139 pKa = 10.4PSSEE143 pKa = 4.13TLGDD147 pKa = 4.54HH148 pKa = 6.76IDD150 pKa = 3.5KK151 pKa = 10.9NGPGDD156 pKa = 3.89YY157 pKa = 10.53EE158 pKa = 5.12SDD160 pKa = 3.27GTTLNDD166 pKa = 4.83DD167 pKa = 4.6DD168 pKa = 5.13PDD170 pKa = 5.62GYY172 pKa = 10.06VTVAFEE178 pKa = 4.16DD179 pKa = 4.62VYY181 pKa = 10.0VTEE184 pKa = 4.5VSFDD188 pKa = 3.58YY189 pKa = 11.28TNGSNAQSDD198 pKa = 4.21PNNQGIYY205 pKa = 9.64LANITFDD212 pKa = 4.6ALDD215 pKa = 3.7SDD217 pKa = 4.47GDD219 pKa = 4.18GIADD223 pKa = 4.09VKK225 pKa = 11.31DD226 pKa = 3.38IDD228 pKa = 5.15DD229 pKa = 5.13DD230 pKa = 4.31NDD232 pKa = 4.58GILDD236 pKa = 3.74VDD238 pKa = 3.93EE239 pKa = 4.55STPAVTEE246 pKa = 4.35SNTASGTIPDD256 pKa = 3.55NGYY259 pKa = 9.82PNTCLDD265 pKa = 3.69RR266 pKa = 11.84TFDD269 pKa = 3.61IQDD272 pKa = 3.3SGIVNDD278 pKa = 3.51VTIKK282 pKa = 10.72VDD284 pKa = 3.86IAHH287 pKa = 6.84TWRR290 pKa = 11.84NDD292 pKa = 3.83LIVQLISPFGTAVDD306 pKa = 5.06LIRR309 pKa = 11.84NEE311 pKa = 4.33GGSHH315 pKa = 6.3NNLSATFFDD324 pKa = 4.23AATTSIVGDD333 pKa = 3.7TTDD336 pKa = 3.59FTLGSFLPRR345 pKa = 11.84QPEE348 pKa = 3.91QALSAFNGEE357 pKa = 4.55DD358 pKa = 3.66PQGTWTLHH366 pKa = 5.13MCDD369 pKa = 5.31DD370 pKa = 4.65ASQDD374 pKa = 2.98TGTFNEE380 pKa = 4.2ANLTINYY387 pKa = 9.36VNEE390 pKa = 4.33LDD392 pKa = 4.13SDD394 pKa = 3.75NDD396 pKa = 4.77GIIDD400 pKa = 4.69CLDD403 pKa = 4.03LDD405 pKa = 4.48SDD407 pKa = 4.52NDD409 pKa = 4.72GIPDD413 pKa = 3.58NVEE416 pKa = 3.93AQPTATYY423 pKa = 10.48DD424 pKa = 3.7EE425 pKa = 5.6PDD427 pKa = 4.04SAWADD432 pKa = 3.26ADD434 pKa = 3.89GDD436 pKa = 4.24GLADD440 pKa = 4.82QYY442 pKa = 11.38DD443 pKa = 4.08TDD445 pKa = 3.85SGGTAVTPSDD455 pKa = 3.74TDD457 pKa = 3.44GDD459 pKa = 4.59GIPDD463 pKa = 3.91YY464 pKa = 11.22LDD466 pKa = 3.81SDD468 pKa = 4.39SDD470 pKa = 3.74NDD472 pKa = 4.29GYY474 pKa = 9.68TDD476 pKa = 3.84CEE478 pKa = 4.31EE479 pKa = 4.53GNSVATGCPITPGDD493 pKa = 3.67TASTKK498 pKa = 10.13TGLVSWAQGSLGDD511 pKa = 4.52VYY513 pKa = 11.04WDD515 pKa = 3.63TTNSASVSNGNVDD528 pKa = 4.8DD529 pKa = 4.84PSNDD533 pKa = 3.31LFNEE537 pKa = 4.22TGDD540 pKa = 3.68TSEE543 pKa = 3.64VGYY546 pKa = 10.99RR547 pKa = 11.84EE548 pKa = 3.96FLCGKK553 pKa = 9.57NRR555 pKa = 11.84TTLTHH560 pKa = 5.86FQWKK564 pKa = 9.23IVSFCCATGSNHH576 pKa = 6.93IEE578 pKa = 4.19DD579 pKa = 5.0LLGGDD584 pKa = 4.21LGAYY588 pKa = 6.49GTDD591 pKa = 2.21WVVFKK596 pKa = 11.02QSGTDD601 pKa = 3.17QYY603 pKa = 11.16EE604 pKa = 4.15INSGHH609 pKa = 6.84KK610 pKa = 9.0NTTKK614 pKa = 10.73AQLAATDD621 pKa = 3.76TVVPGKK627 pKa = 10.3GYY629 pKa = 9.86WIIADD634 pKa = 3.94LGGAGNEE641 pKa = 4.09KK642 pKa = 10.71NITIDD647 pKa = 3.22KK648 pKa = 9.06TLSNLSPASTVTSSSVGITNPDD670 pKa = 3.01FTEE673 pKa = 3.81VHH675 pKa = 6.68EE676 pKa = 4.27YY677 pKa = 10.89LLPKK681 pKa = 10.7NEE683 pKa = 3.92VSDD686 pKa = 4.66PNTVDD691 pKa = 3.16YY692 pKa = 10.68KK693 pKa = 11.22KK694 pKa = 11.27YY695 pKa = 9.01MAGNPFPYY703 pKa = 10.29AFQLSDD709 pKa = 4.87LYY711 pKa = 10.94FKK713 pKa = 11.07HH714 pKa = 6.28NAGGMSYY721 pKa = 11.3NEE723 pKa = 4.53MGNTANDD730 pKa = 3.19NYY732 pKa = 10.27INKK735 pKa = 9.18IVYY738 pKa = 9.71KK739 pKa = 10.25HH740 pKa = 6.83DD741 pKa = 4.18SNKK744 pKa = 9.25TGPVSGYY751 pKa = 9.09MAVDD755 pKa = 3.8PATPGFDD762 pKa = 4.17GSIQPMEE769 pKa = 4.04GFFIKK774 pKa = 9.94IEE776 pKa = 4.03KK777 pKa = 9.66NQTDD781 pKa = 3.38NYY783 pKa = 9.06VNHH786 pKa = 6.65FAYY789 pKa = 10.0PLMNKK794 pKa = 9.64
Molecular weight: 85.68 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|A6QAM9|DAPD_SULNB 2 3 4 5-tetrahydropyridine-2 6-dicarboxylate N-succinyltransferase OS=Sulfurovum sp. (strain NBC37-1) OX=387093 GN=dapD PE=3 SV=1
MM1 pKa = 7.28KK2 pKa = 9.44RR3 pKa = 11.84TYY5 pKa = 10.04QPHH8 pKa = 4.86NTPRR12 pKa = 11.84KK13 pKa = 7.34RR14 pKa = 11.84THH16 pKa = 6.22GFRR19 pKa = 11.84TRR21 pKa = 11.84MKK23 pKa = 8.87TKK25 pKa = 10.11NGRR28 pKa = 11.84KK29 pKa = 9.33VISARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.83GRR39 pKa = 11.84KK40 pKa = 8.68RR41 pKa = 11.84LSVV44 pKa = 3.2
MM1 pKa = 7.28KK2 pKa = 9.44RR3 pKa = 11.84TYY5 pKa = 10.04QPHH8 pKa = 4.86NTPRR12 pKa = 11.84KK13 pKa = 7.34RR14 pKa = 11.84THH16 pKa = 6.22GFRR19 pKa = 11.84TRR21 pKa = 11.84MKK23 pKa = 8.87TKK25 pKa = 10.11NGRR28 pKa = 11.84KK29 pKa = 9.33VISARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.83GRR39 pKa = 11.84KK40 pKa = 8.68RR41 pKa = 11.84LSVV44 pKa = 3.2
Molecular weight: 5.29 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
765727 |
37 |
2052 |
314.7 |
35.5 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.369 ± 0.049 | 0.867 ± 0.015 |
5.628 ± 0.038 | 7.189 ± 0.046 |
4.713 ± 0.035 | 6.413 ± 0.048 |
2.193 ± 0.025 | 7.652 ± 0.045 |
8.08 ± 0.061 | 9.686 ± 0.056 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.896 ± 0.026 | 4.259 ± 0.041 |
3.45 ± 0.027 | 3.007 ± 0.026 |
3.983 ± 0.037 | 5.928 ± 0.04 |
5.258 ± 0.038 | 6.499 ± 0.046 |
0.95 ± 0.015 | 3.979 ± 0.033 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
