
Nostoc sp. (strain ATCC 29411 / PCC 7524)
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Cyanobacteria/Melainabacteria group; Cyanobacteria; Nostocales; Nostocaceae; Nostoc; unclassified Nostoc
Average proteome isoelectric point is 6.3
Get precalculated fractions of proteins
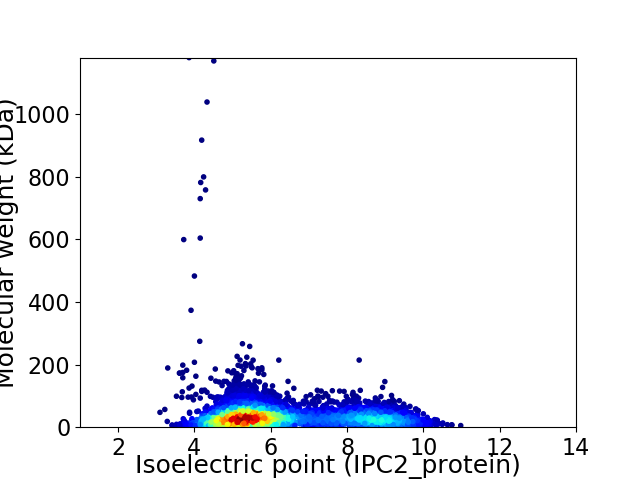
Virtual 2D-PAGE plot for 5398 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|K9QX42|K9QX42_NOSS7 Anti-sigma factor antagonist OS=Nostoc sp. (strain ATCC 29411 / PCC 7524) OX=28072 GN=Nos7524_4237 PE=3 SV=1
MM1 pKa = 6.37TTYY4 pKa = 10.74SGFPDD9 pKa = 4.28PGSSTSTALDD19 pKa = 3.74TTSLSYY25 pKa = 10.56IGGSLLSTPSIFQDD39 pKa = 3.36SVSRR43 pKa = 11.84LSPSDD48 pKa = 3.24LSDD51 pKa = 3.39YY52 pKa = 11.35YY53 pKa = 11.1RR54 pKa = 11.84FTVSSSSIVTLEE66 pKa = 4.73LDD68 pKa = 4.0GLTDD72 pKa = 3.53NANLFLRR79 pKa = 11.84NSAGSSIAPSTNAGTTAEE97 pKa = 4.65RR98 pKa = 11.84IRR100 pKa = 11.84FSISDD105 pKa = 3.64PPSNPFYY112 pKa = 11.05AHH114 pKa = 6.07VFIPNGSTSTPYY126 pKa = 10.99NLRR129 pKa = 11.84LSAEE133 pKa = 4.66TIQEE137 pKa = 4.21SYY139 pKa = 10.89ISDD142 pKa = 3.67SLIGNSRR149 pKa = 11.84NIGTFSTTDD158 pKa = 4.11GITPIIDD165 pKa = 3.69YY166 pKa = 10.61VSSTGQVVDD175 pKa = 3.46QNDD178 pKa = 4.31YY179 pKa = 11.72YY180 pKa = 11.77SFTLSNSGTVDD191 pKa = 3.02INLSSLNGTDD201 pKa = 3.46ILYY204 pKa = 10.7ADD206 pKa = 4.37LQLINSFNSVLQTSANVGTTSEE228 pKa = 4.63LINYY232 pKa = 8.7SLAAGTYY239 pKa = 8.07YY240 pKa = 9.96IRR242 pKa = 11.84AYY244 pKa = 10.69SLSDD248 pKa = 3.27PGNYY252 pKa = 9.21QLNFNFTADD261 pKa = 4.01PPDD264 pKa = 3.24QGGNTVGTATPIILDD279 pKa = 3.66TTITDD284 pKa = 3.77QVSLADD290 pKa = 3.51NSDD293 pKa = 3.89YY294 pKa = 11.5YY295 pKa = 11.44QFTLASTSLVDD306 pKa = 4.48IRR308 pKa = 11.84FTSLTADD315 pKa = 3.32ANLFLQNASGGNIITSTQPGTALDD339 pKa = 3.89AVRR342 pKa = 11.84LSLNPGTYY350 pKa = 10.24NILVNRR356 pKa = 11.84GSTQTAEE363 pKa = 4.26YY364 pKa = 8.77TLSASAQVIGTDD376 pKa = 3.3QAPNSTAIALNLGNIIGATSRR397 pKa = 11.84NEE399 pKa = 3.85FVGNLDD405 pKa = 3.36TNDD408 pKa = 3.54FYY410 pKa = 11.82KK411 pKa = 10.19FTLDD415 pKa = 3.0TTTNFSLNLNILSSYY430 pKa = 10.96FNAQLVNADD439 pKa = 3.71VQLLTSNGTQIAISNQPGNANEE461 pKa = 4.78SINTTLDD468 pKa = 2.79AGTYY472 pKa = 8.88FIRR475 pKa = 11.84VYY477 pKa = 8.51TTGLANTFYY486 pKa = 11.06DD487 pKa = 4.36LDD489 pKa = 3.66ITATPQAKK497 pKa = 9.5LVQNINPTGSSNPANLTALGSLVYY521 pKa = 8.09FTANDD526 pKa = 3.93GVNGVQLWRR535 pKa = 11.84SDD537 pKa = 3.38GNTATSLSNISSFNPNNLVVFNNRR561 pKa = 11.84LYY563 pKa = 9.88FTASDD568 pKa = 3.63STFGRR573 pKa = 11.84EE574 pKa = 3.32LWEE577 pKa = 4.09YY578 pKa = 11.23NGTSVNRR585 pKa = 11.84ISDD588 pKa = 3.71INVGAGNSDD597 pKa = 3.66PGNLTVVGNKK607 pKa = 10.01LFFTAVDD614 pKa = 3.31SSTTRR619 pKa = 11.84KK620 pKa = 9.23LWVYY624 pKa = 10.93NGTSVNLVDD633 pKa = 4.56INPGFATTGTPTFTTAFNNRR653 pKa = 11.84LFFTTQNNSQLWSTDD668 pKa = 2.67GTVGGTQVINAGGVTNSTPRR688 pKa = 11.84SLTVVGNTLYY698 pKa = 9.09FTANNGTSGHH708 pKa = 5.92EE709 pKa = 3.78VWQYY713 pKa = 11.33QSGTTASLVEE723 pKa = 5.67DD724 pKa = 3.74ITPGNNSYY732 pKa = 11.13APEE735 pKa = 4.4RR736 pKa = 11.84LTTVGNMLYY745 pKa = 10.44FVTDD749 pKa = 3.24SDD751 pKa = 4.23NDD753 pKa = 3.51FTLEE757 pKa = 3.93LWRR760 pKa = 11.84SDD762 pKa = 3.09GTEE765 pKa = 3.81SGTQIIGTDD774 pKa = 3.47GQAPNLGFGAINLTAVGNTLYY795 pKa = 10.51FVANDD800 pKa = 3.78PVSGLEE806 pKa = 3.75LWKK809 pKa = 9.73TDD811 pKa = 3.13GTDD814 pKa = 2.86VGTVIVKK821 pKa = 10.32DD822 pKa = 3.68ISSGTANSLPTGLVNFNGTLAFAASDD848 pKa = 3.59GTNRR852 pKa = 11.84EE853 pKa = 3.77VWFSDD858 pKa = 2.96GTEE861 pKa = 4.17LNTIRR866 pKa = 11.84ASNIYY871 pKa = 9.56PGGIANPDD879 pKa = 3.18WLTVVGTKK887 pKa = 10.62LFFTATDD894 pKa = 3.8GADD897 pKa = 3.36NTEE900 pKa = 4.37LYY902 pKa = 10.92VLL904 pKa = 4.45
MM1 pKa = 6.37TTYY4 pKa = 10.74SGFPDD9 pKa = 4.28PGSSTSTALDD19 pKa = 3.74TTSLSYY25 pKa = 10.56IGGSLLSTPSIFQDD39 pKa = 3.36SVSRR43 pKa = 11.84LSPSDD48 pKa = 3.24LSDD51 pKa = 3.39YY52 pKa = 11.35YY53 pKa = 11.1RR54 pKa = 11.84FTVSSSSIVTLEE66 pKa = 4.73LDD68 pKa = 4.0GLTDD72 pKa = 3.53NANLFLRR79 pKa = 11.84NSAGSSIAPSTNAGTTAEE97 pKa = 4.65RR98 pKa = 11.84IRR100 pKa = 11.84FSISDD105 pKa = 3.64PPSNPFYY112 pKa = 11.05AHH114 pKa = 6.07VFIPNGSTSTPYY126 pKa = 10.99NLRR129 pKa = 11.84LSAEE133 pKa = 4.66TIQEE137 pKa = 4.21SYY139 pKa = 10.89ISDD142 pKa = 3.67SLIGNSRR149 pKa = 11.84NIGTFSTTDD158 pKa = 4.11GITPIIDD165 pKa = 3.69YY166 pKa = 10.61VSSTGQVVDD175 pKa = 3.46QNDD178 pKa = 4.31YY179 pKa = 11.72YY180 pKa = 11.77SFTLSNSGTVDD191 pKa = 3.02INLSSLNGTDD201 pKa = 3.46ILYY204 pKa = 10.7ADD206 pKa = 4.37LQLINSFNSVLQTSANVGTTSEE228 pKa = 4.63LINYY232 pKa = 8.7SLAAGTYY239 pKa = 8.07YY240 pKa = 9.96IRR242 pKa = 11.84AYY244 pKa = 10.69SLSDD248 pKa = 3.27PGNYY252 pKa = 9.21QLNFNFTADD261 pKa = 4.01PPDD264 pKa = 3.24QGGNTVGTATPIILDD279 pKa = 3.66TTITDD284 pKa = 3.77QVSLADD290 pKa = 3.51NSDD293 pKa = 3.89YY294 pKa = 11.5YY295 pKa = 11.44QFTLASTSLVDD306 pKa = 4.48IRR308 pKa = 11.84FTSLTADD315 pKa = 3.32ANLFLQNASGGNIITSTQPGTALDD339 pKa = 3.89AVRR342 pKa = 11.84LSLNPGTYY350 pKa = 10.24NILVNRR356 pKa = 11.84GSTQTAEE363 pKa = 4.26YY364 pKa = 8.77TLSASAQVIGTDD376 pKa = 3.3QAPNSTAIALNLGNIIGATSRR397 pKa = 11.84NEE399 pKa = 3.85FVGNLDD405 pKa = 3.36TNDD408 pKa = 3.54FYY410 pKa = 11.82KK411 pKa = 10.19FTLDD415 pKa = 3.0TTTNFSLNLNILSSYY430 pKa = 10.96FNAQLVNADD439 pKa = 3.71VQLLTSNGTQIAISNQPGNANEE461 pKa = 4.78SINTTLDD468 pKa = 2.79AGTYY472 pKa = 8.88FIRR475 pKa = 11.84VYY477 pKa = 8.51TTGLANTFYY486 pKa = 11.06DD487 pKa = 4.36LDD489 pKa = 3.66ITATPQAKK497 pKa = 9.5LVQNINPTGSSNPANLTALGSLVYY521 pKa = 8.09FTANDD526 pKa = 3.93GVNGVQLWRR535 pKa = 11.84SDD537 pKa = 3.38GNTATSLSNISSFNPNNLVVFNNRR561 pKa = 11.84LYY563 pKa = 9.88FTASDD568 pKa = 3.63STFGRR573 pKa = 11.84EE574 pKa = 3.32LWEE577 pKa = 4.09YY578 pKa = 11.23NGTSVNRR585 pKa = 11.84ISDD588 pKa = 3.71INVGAGNSDD597 pKa = 3.66PGNLTVVGNKK607 pKa = 10.01LFFTAVDD614 pKa = 3.31SSTTRR619 pKa = 11.84KK620 pKa = 9.23LWVYY624 pKa = 10.93NGTSVNLVDD633 pKa = 4.56INPGFATTGTPTFTTAFNNRR653 pKa = 11.84LFFTTQNNSQLWSTDD668 pKa = 2.67GTVGGTQVINAGGVTNSTPRR688 pKa = 11.84SLTVVGNTLYY698 pKa = 9.09FTANNGTSGHH708 pKa = 5.92EE709 pKa = 3.78VWQYY713 pKa = 11.33QSGTTASLVEE723 pKa = 5.67DD724 pKa = 3.74ITPGNNSYY732 pKa = 11.13APEE735 pKa = 4.4RR736 pKa = 11.84LTTVGNMLYY745 pKa = 10.44FVTDD749 pKa = 3.24SDD751 pKa = 4.23NDD753 pKa = 3.51FTLEE757 pKa = 3.93LWRR760 pKa = 11.84SDD762 pKa = 3.09GTEE765 pKa = 3.81SGTQIIGTDD774 pKa = 3.47GQAPNLGFGAINLTAVGNTLYY795 pKa = 10.51FVANDD800 pKa = 3.78PVSGLEE806 pKa = 3.75LWKK809 pKa = 9.73TDD811 pKa = 3.13GTDD814 pKa = 2.86VGTVIVKK821 pKa = 10.32DD822 pKa = 3.68ISSGTANSLPTGLVNFNGTLAFAASDD848 pKa = 3.59GTNRR852 pKa = 11.84EE853 pKa = 3.77VWFSDD858 pKa = 2.96GTEE861 pKa = 4.17LNTIRR866 pKa = 11.84ASNIYY871 pKa = 9.56PGGIANPDD879 pKa = 3.18WLTVVGTKK887 pKa = 10.62LFFTATDD894 pKa = 3.8GADD897 pKa = 3.36NTEE900 pKa = 4.37LYY902 pKa = 10.92VLL904 pKa = 4.45
Molecular weight: 96.14 kDa
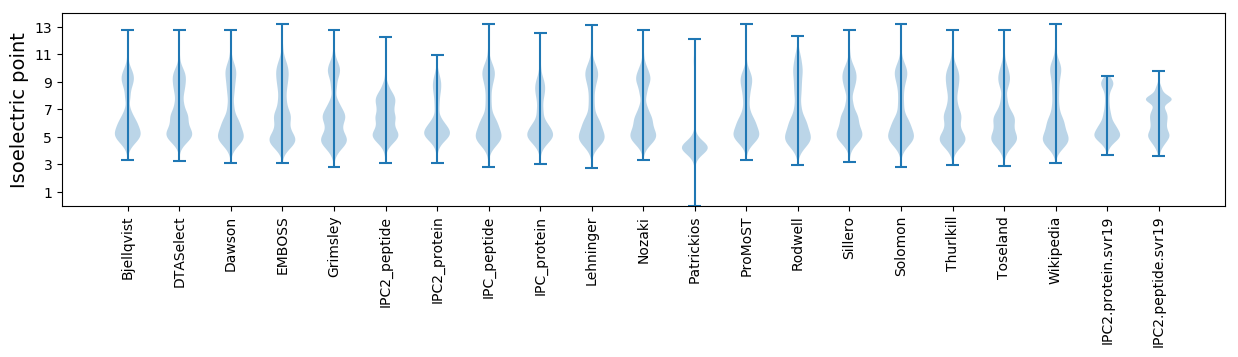
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|K9QUY1|K9QUY1_NOSS7 Phosphate acyltransferase OS=Nostoc sp. (strain ATCC 29411 / PCC 7524) OX=28072 GN=plsX PE=3 SV=1
MM1 pKa = 7.08QRR3 pKa = 11.84TLGGTNRR10 pKa = 11.84KK11 pKa = 9.1RR12 pKa = 11.84KK13 pKa = 7.65RR14 pKa = 11.84TSGFRR19 pKa = 11.84ARR21 pKa = 11.84MRR23 pKa = 11.84TPDD26 pKa = 3.17GRR28 pKa = 11.84NVIRR32 pKa = 11.84ARR34 pKa = 11.84RR35 pKa = 11.84KK36 pKa = 9.01KK37 pKa = 9.6GRR39 pKa = 11.84HH40 pKa = 5.0RR41 pKa = 11.84LSVV44 pKa = 3.12
MM1 pKa = 7.08QRR3 pKa = 11.84TLGGTNRR10 pKa = 11.84KK11 pKa = 9.1RR12 pKa = 11.84KK13 pKa = 7.65RR14 pKa = 11.84TSGFRR19 pKa = 11.84ARR21 pKa = 11.84MRR23 pKa = 11.84TPDD26 pKa = 3.17GRR28 pKa = 11.84NVIRR32 pKa = 11.84ARR34 pKa = 11.84RR35 pKa = 11.84KK36 pKa = 9.01KK37 pKa = 9.6GRR39 pKa = 11.84HH40 pKa = 5.0RR41 pKa = 11.84LSVV44 pKa = 3.12
Molecular weight: 5.22 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1809286 |
29 |
11414 |
335.2 |
37.25 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.217 ± 0.045 | 0.929 ± 0.018 |
4.934 ± 0.045 | 5.975 ± 0.048 |
3.847 ± 0.026 | 6.813 ± 0.073 |
1.857 ± 0.025 | 6.913 ± 0.026 |
4.481 ± 0.044 | 10.802 ± 0.065 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.779 ± 0.024 | 4.667 ± 0.064 |
4.717 ± 0.042 | 5.58 ± 0.048 |
4.947 ± 0.04 | 6.277 ± 0.04 |
6.072 ± 0.077 | 6.723 ± 0.031 |
1.401 ± 0.021 | 3.07 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
