
TM7 phylum sp. oral taxon 356
Taxonomy: cellular organisms; Bacteria; Bacteria incertae sedis; Bacteria candidate phyla; Candidatus Saccharibacteria
Average proteome isoelectric point is 6.96
Get precalculated fractions of proteins
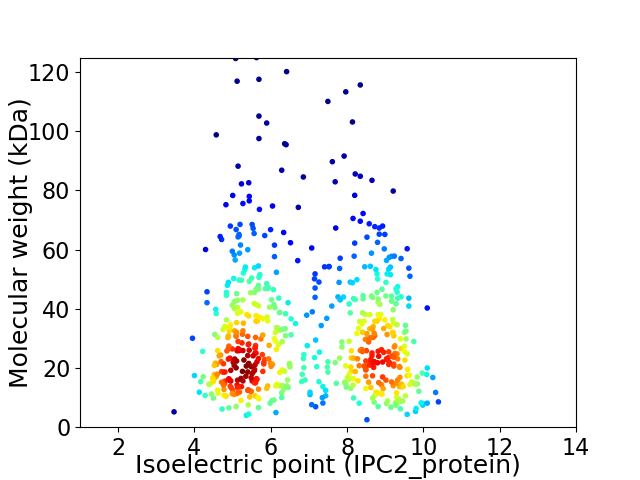
Virtual 2D-PAGE plot for 611 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A563AES4|A0A563AES4_9BACT Phage holin family protein OS=TM7 phylum sp. oral taxon 356 OX=713057 GN=EUA81_01605 PE=4 SV=1
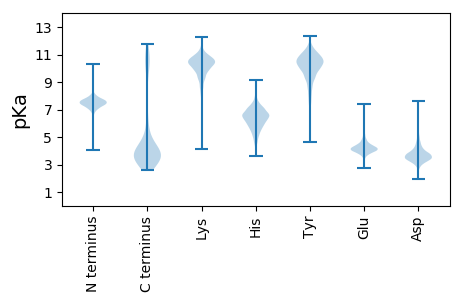
MM1 pKa = 7.92HH2 pKa = 6.91YY3 pKa = 10.49NQYY6 pKa = 10.2QRR8 pKa = 11.84LINIVGGLYY17 pKa = 10.22EE18 pKa = 3.93NHH20 pKa = 6.83LGYY23 pKa = 10.4FDD25 pKa = 5.94DD26 pKa = 4.14LTAEE30 pKa = 4.17EE31 pKa = 4.44RR32 pKa = 11.84QVLSRR37 pKa = 11.84VFFYY41 pKa = 10.56DD42 pKa = 3.2YY43 pKa = 10.81DD44 pKa = 4.39YY45 pKa = 11.67DD46 pKa = 5.76SEE48 pKa = 4.9DD49 pKa = 4.32CPDD52 pKa = 4.66DD53 pKa = 4.18FPEE56 pKa = 4.26SFPDD60 pKa = 4.05FFRR63 pKa = 11.84DD64 pKa = 4.54RR65 pKa = 11.84IAGNQALQDD74 pKa = 3.58EE75 pKa = 4.86ALAAVARR82 pKa = 11.84LYY84 pKa = 11.45AMSGMGDD91 pKa = 3.74FALTRR96 pKa = 11.84VSDD99 pKa = 3.68KK100 pKa = 11.03PLL102 pKa = 4.58
MM1 pKa = 7.92HH2 pKa = 6.91YY3 pKa = 10.49NQYY6 pKa = 10.2QRR8 pKa = 11.84LINIVGGLYY17 pKa = 10.22EE18 pKa = 3.93NHH20 pKa = 6.83LGYY23 pKa = 10.4FDD25 pKa = 5.94DD26 pKa = 4.14LTAEE30 pKa = 4.17EE31 pKa = 4.44RR32 pKa = 11.84QVLSRR37 pKa = 11.84VFFYY41 pKa = 10.56DD42 pKa = 3.2YY43 pKa = 10.81DD44 pKa = 4.39YY45 pKa = 11.67DD46 pKa = 5.76SEE48 pKa = 4.9DD49 pKa = 4.32CPDD52 pKa = 4.66DD53 pKa = 4.18FPEE56 pKa = 4.26SFPDD60 pKa = 4.05FFRR63 pKa = 11.84DD64 pKa = 4.54RR65 pKa = 11.84IAGNQALQDD74 pKa = 3.58EE75 pKa = 4.86ALAAVARR82 pKa = 11.84LYY84 pKa = 11.45AMSGMGDD91 pKa = 3.74FALTRR96 pKa = 11.84VSDD99 pKa = 3.68KK100 pKa = 11.03PLL102 pKa = 4.58
Molecular weight: 11.84 kDa
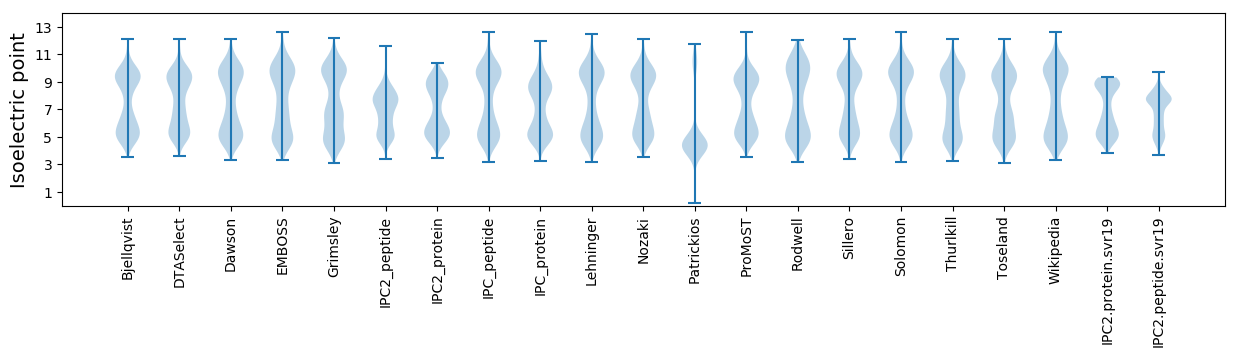
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A563ADB0|A0A563ADB0_9BACT Response regulator OS=TM7 phylum sp. oral taxon 356 OX=713057 GN=EUA81_02960 PE=4 SV=1
MM1 pKa = 7.04YY2 pKa = 10.13KK3 pKa = 10.19VNRR6 pKa = 11.84AMEE9 pKa = 4.3RR10 pKa = 11.84WRR12 pKa = 11.84YY13 pKa = 7.49NANRR17 pKa = 11.84VSAGGSSWAGRR28 pKa = 11.84VDD30 pKa = 3.65PAKK33 pKa = 10.59SRR35 pKa = 11.84QNQITARR42 pKa = 11.84QIKK45 pKa = 10.57ADD47 pKa = 3.75LFDD50 pKa = 6.13DD51 pKa = 4.18NGEE54 pKa = 3.97LKK56 pKa = 10.48LARR59 pKa = 11.84RR60 pKa = 11.84HH61 pKa = 5.71RR62 pKa = 11.84ADD64 pKa = 3.15HH65 pKa = 6.62GIVALAVALSIFGVVIIYY83 pKa = 10.27SIASGLYY90 pKa = 9.25GGNTTVINQEE100 pKa = 3.79MLKK103 pKa = 10.41RR104 pKa = 11.84GIFLAVGVVLFIVASRR120 pKa = 11.84IPLNWWKK127 pKa = 10.7KK128 pKa = 7.63AGPYY132 pKa = 9.89IFLIALLICLALPILGAAHH151 pKa = 6.15VPVARR156 pKa = 11.84CALGACRR163 pKa = 11.84WYY165 pKa = 10.88KK166 pKa = 10.69LGVVSFQPAEE176 pKa = 3.93FLKK179 pKa = 10.99LGTVLFIAGYY189 pKa = 10.4LSTRR193 pKa = 11.84LANGKK198 pKa = 10.06LNDD201 pKa = 4.04KK202 pKa = 9.34LTLAEE207 pKa = 4.22VLAVMLVSLLIIVGLQRR224 pKa = 11.84DD225 pKa = 4.21MGTGAAIVAIFLIEE239 pKa = 3.93LVMSGMSWRR248 pKa = 11.84KK249 pKa = 8.75LAIVLGVIVLLFVVMTLAAPHH270 pKa = 5.86RR271 pKa = 11.84MSRR274 pKa = 11.84IFTFTKK280 pKa = 10.14SGSNADD286 pKa = 3.07SDD288 pKa = 4.08YY289 pKa = 11.06HH290 pKa = 8.11INQALIGLGSGGLTGRR306 pKa = 11.84GLGQSVQAFGWLPEE320 pKa = 4.11AVNDD324 pKa = 4.5SIFAIVGEE332 pKa = 4.24TLGFVGTVGLIATFAFLIMRR352 pKa = 11.84MFSKK356 pKa = 9.91TNYY359 pKa = 8.95LNNMYY364 pKa = 10.36LRR366 pKa = 11.84LVVAGVVGWLASHH379 pKa = 6.61VLANVMAMTHH389 pKa = 7.11LIPLTGITLPLVSSGGTSAIFVMISLGVVFEE420 pKa = 4.19ISCYY424 pKa = 7.0TAHH427 pKa = 6.9RR428 pKa = 11.84RR429 pKa = 11.84IDD431 pKa = 3.2IAEE434 pKa = 4.13VNEE437 pKa = 4.31SQGGDD442 pKa = 3.27SEE444 pKa = 4.84NPVRR448 pKa = 11.84RR449 pKa = 11.84RR450 pKa = 11.84RR451 pKa = 11.84QRR453 pKa = 11.84WSHH456 pKa = 4.51STDD459 pKa = 2.61RR460 pKa = 11.84RR461 pKa = 11.84RR462 pKa = 11.84SRR464 pKa = 11.84TTTSAGG470 pKa = 3.27
MM1 pKa = 7.04YY2 pKa = 10.13KK3 pKa = 10.19VNRR6 pKa = 11.84AMEE9 pKa = 4.3RR10 pKa = 11.84WRR12 pKa = 11.84YY13 pKa = 7.49NANRR17 pKa = 11.84VSAGGSSWAGRR28 pKa = 11.84VDD30 pKa = 3.65PAKK33 pKa = 10.59SRR35 pKa = 11.84QNQITARR42 pKa = 11.84QIKK45 pKa = 10.57ADD47 pKa = 3.75LFDD50 pKa = 6.13DD51 pKa = 4.18NGEE54 pKa = 3.97LKK56 pKa = 10.48LARR59 pKa = 11.84RR60 pKa = 11.84HH61 pKa = 5.71RR62 pKa = 11.84ADD64 pKa = 3.15HH65 pKa = 6.62GIVALAVALSIFGVVIIYY83 pKa = 10.27SIASGLYY90 pKa = 9.25GGNTTVINQEE100 pKa = 3.79MLKK103 pKa = 10.41RR104 pKa = 11.84GIFLAVGVVLFIVASRR120 pKa = 11.84IPLNWWKK127 pKa = 10.7KK128 pKa = 7.63AGPYY132 pKa = 9.89IFLIALLICLALPILGAAHH151 pKa = 6.15VPVARR156 pKa = 11.84CALGACRR163 pKa = 11.84WYY165 pKa = 10.88KK166 pKa = 10.69LGVVSFQPAEE176 pKa = 3.93FLKK179 pKa = 10.99LGTVLFIAGYY189 pKa = 10.4LSTRR193 pKa = 11.84LANGKK198 pKa = 10.06LNDD201 pKa = 4.04KK202 pKa = 9.34LTLAEE207 pKa = 4.22VLAVMLVSLLIIVGLQRR224 pKa = 11.84DD225 pKa = 4.21MGTGAAIVAIFLIEE239 pKa = 3.93LVMSGMSWRR248 pKa = 11.84KK249 pKa = 8.75LAIVLGVIVLLFVVMTLAAPHH270 pKa = 5.86RR271 pKa = 11.84MSRR274 pKa = 11.84IFTFTKK280 pKa = 10.14SGSNADD286 pKa = 3.07SDD288 pKa = 4.08YY289 pKa = 11.06HH290 pKa = 8.11INQALIGLGSGGLTGRR306 pKa = 11.84GLGQSVQAFGWLPEE320 pKa = 4.11AVNDD324 pKa = 4.5SIFAIVGEE332 pKa = 4.24TLGFVGTVGLIATFAFLIMRR352 pKa = 11.84MFSKK356 pKa = 9.91TNYY359 pKa = 8.95LNNMYY364 pKa = 10.36LRR366 pKa = 11.84LVVAGVVGWLASHH379 pKa = 6.61VLANVMAMTHH389 pKa = 7.11LIPLTGITLPLVSSGGTSAIFVMISLGVVFEE420 pKa = 4.19ISCYY424 pKa = 7.0TAHH427 pKa = 6.9RR428 pKa = 11.84RR429 pKa = 11.84IDD431 pKa = 3.2IAEE434 pKa = 4.13VNEE437 pKa = 4.31SQGGDD442 pKa = 3.27SEE444 pKa = 4.84NPVRR448 pKa = 11.84RR449 pKa = 11.84RR450 pKa = 11.84RR451 pKa = 11.84QRR453 pKa = 11.84WSHH456 pKa = 4.51STDD459 pKa = 2.61RR460 pKa = 11.84RR461 pKa = 11.84RR462 pKa = 11.84SRR464 pKa = 11.84TTTSAGG470 pKa = 3.27
Molecular weight: 51.02 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
181644 |
25 |
1106 |
297.3 |
33.13 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.442 ± 0.104 | 0.747 ± 0.026 |
6.175 ± 0.094 | 5.518 ± 0.096 |
3.47 ± 0.064 | 6.732 ± 0.092 |
1.915 ± 0.048 | 7.115 ± 0.099 |
6.032 ± 0.087 | 9.328 ± 0.112 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.311 ± 0.041 | 4.421 ± 0.075 |
3.994 ± 0.075 | 4.167 ± 0.077 |
5.757 ± 0.075 | 6.829 ± 0.089 |
5.576 ± 0.074 | 6.84 ± 0.09 |
1.056 ± 0.032 | 3.577 ± 0.064 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
