
Hubei sobemo-like virus 2
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 6.98
Get precalculated fractions of proteins
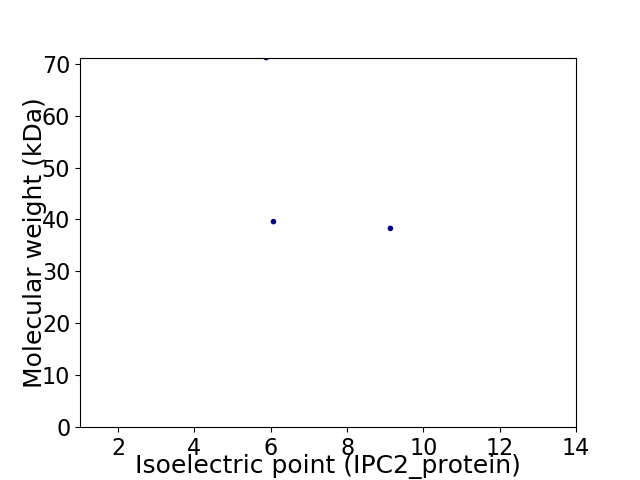
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KEZ9|A0A1L3KEZ9_9VIRU Peptidase S39 domain-containing protein OS=Hubei sobemo-like virus 2 OX=1923205 PE=4 SV=1
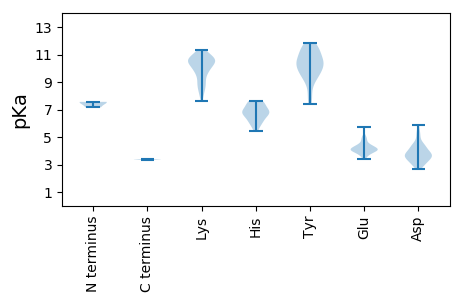
MM1 pKa = 7.16MWTATLEE8 pKa = 4.01NSAIASRR15 pKa = 11.84QLQLSAVEE23 pKa = 3.95LWEE26 pKa = 4.98DD27 pKa = 3.41YY28 pKa = 11.17SYY30 pKa = 11.7LLNYY34 pKa = 9.55FFLLGVASVLAVVAIRR50 pKa = 11.84AAVNYY55 pKa = 10.07YY56 pKa = 10.61EE57 pKa = 4.31EE58 pKa = 4.98LRR60 pKa = 11.84LDD62 pKa = 3.61SLVRR66 pKa = 11.84EE67 pKa = 4.57LPTPPEE73 pKa = 4.25DD74 pKa = 3.53PVLDD78 pKa = 3.76GHH80 pKa = 7.62LEE82 pKa = 3.98FCPNNGYY89 pKa = 8.99VMRR92 pKa = 11.84VTTVIDD98 pKa = 4.0DD99 pKa = 3.99EE100 pKa = 4.42QHH102 pKa = 5.79SFLVKK107 pKa = 8.97VTPSQVTDD115 pKa = 3.47MLQGSSKK122 pKa = 8.06STRR125 pKa = 11.84VKK127 pKa = 10.05EE128 pKa = 4.07ARR130 pKa = 11.84GSSGVVPPGPRR141 pKa = 11.84AWDD144 pKa = 3.76AGNSTVPPAVHH155 pKa = 6.72LNGIPSAEE163 pKa = 3.87EE164 pKa = 3.43HH165 pKa = 7.18DD166 pKa = 3.66FDD168 pKa = 5.52LRR170 pKa = 11.84VVRR173 pKa = 11.84ALVRR177 pKa = 11.84MGVEE181 pKa = 3.7TRR183 pKa = 11.84EE184 pKa = 4.09NIQALGWDD192 pKa = 4.21FPEE195 pKa = 4.56ITASDD200 pKa = 4.03PQTPTTEE207 pKa = 4.04RR208 pKa = 11.84LLSEE212 pKa = 4.11TATSQEE218 pKa = 4.24LEE220 pKa = 4.11MAQPSSVVVPAQEE233 pKa = 4.46AFTKK237 pKa = 10.38KK238 pKa = 9.76ILKK241 pKa = 8.47ITDD244 pKa = 3.17RR245 pKa = 11.84EE246 pKa = 4.51GNVRR250 pKa = 11.84GLGFRR255 pKa = 11.84WEE257 pKa = 4.37NYY259 pKa = 9.72LVTAYY264 pKa = 10.05HH265 pKa = 6.34VYY267 pKa = 10.58EE268 pKa = 4.11SVKK271 pKa = 10.9NEE273 pKa = 3.92EE274 pKa = 4.73CDD276 pKa = 3.63VSPVPSTQGFPLSQFSALTYY296 pKa = 10.4SPKK299 pKa = 10.58LDD301 pKa = 4.29FIVLKK306 pKa = 10.37VDD308 pKa = 3.12KK309 pKa = 10.83PEE311 pKa = 4.08MVWAKK316 pKa = 10.39IGVSKK321 pKa = 10.64MSLKK325 pKa = 9.83TPAPNTPFKK334 pKa = 11.33VIGKK338 pKa = 8.9QNNSWNSSTGFLRR351 pKa = 11.84AAHH354 pKa = 6.79GDD356 pKa = 3.58LKK358 pKa = 10.57IKK360 pKa = 10.78HH361 pKa = 6.2NATTTKK367 pKa = 10.1GYY369 pKa = 10.2SGSPLLSDD377 pKa = 3.66GKK379 pKa = 11.15VVGLHH384 pKa = 6.62LGAANPTNYY393 pKa = 10.37CLDD396 pKa = 3.73LSFLPLLFQEE406 pKa = 5.05GVEE409 pKa = 4.24SEE411 pKa = 4.38EE412 pKa = 4.01KK413 pKa = 9.51QGRR416 pKa = 11.84YY417 pKa = 9.21HH418 pKa = 6.85KK419 pKa = 10.59FVDD422 pKa = 3.2SRR424 pKa = 11.84EE425 pKa = 4.27YY426 pKa = 10.33QDD428 pKa = 4.36HH429 pKa = 7.44ADD431 pKa = 5.2FKK433 pKa = 11.35VEE435 pKa = 3.41WEE437 pKa = 4.01EE438 pKa = 3.77RR439 pKa = 11.84RR440 pKa = 11.84KK441 pKa = 10.42EE442 pKa = 4.01EE443 pKa = 4.11RR444 pKa = 11.84DD445 pKa = 2.97EE446 pKa = 4.88DD447 pKa = 3.75YY448 pKa = 10.96TYY450 pKa = 11.85GLVHH454 pKa = 6.83NKK456 pKa = 9.69RR457 pKa = 11.84FIEE460 pKa = 3.65MRR462 pKa = 11.84YY463 pKa = 9.43RR464 pKa = 11.84KK465 pKa = 10.01SFGDD469 pKa = 3.42ATIVNASEE477 pKa = 3.69EE478 pKa = 4.24TTRR481 pKa = 11.84NFFNNKK487 pKa = 9.04ALPAGVRR494 pKa = 11.84WADD497 pKa = 3.28EE498 pKa = 4.07DD499 pKa = 4.66DD500 pKa = 4.49YY501 pKa = 11.46EE502 pKa = 5.75AVMPDD507 pKa = 3.94FVHH510 pKa = 7.56ASKK513 pKa = 10.78KK514 pKa = 7.96PQLRR518 pKa = 11.84GTEE521 pKa = 4.44TTCGMTQKK529 pKa = 10.8SSGPPPLVTSAPASVSSSSEE549 pKa = 3.75KK550 pKa = 10.62AKK552 pKa = 10.6PSAKK556 pKa = 8.48PTSPPTSILKK566 pKa = 9.61PSPKK570 pKa = 10.01LSSNDD575 pKa = 3.37QSSKK579 pKa = 9.48PSVVFKK585 pKa = 10.5EE586 pKa = 4.13VEE588 pKa = 4.02MQKK591 pKa = 10.56NSPSTSKK598 pKa = 10.01PASSGQPKK606 pKa = 9.73SRR608 pKa = 11.84KK609 pKa = 8.62SKK611 pKa = 10.56KK612 pKa = 9.58KK613 pKa = 8.3EE614 pKa = 3.45HH615 pKa = 6.14SQAFSAIIQNLEE627 pKa = 4.0SLSDD631 pKa = 3.74SEE633 pKa = 5.11VRR635 pKa = 11.84DD636 pKa = 3.46LKK638 pKa = 11.34SRR640 pKa = 11.84IGG642 pKa = 3.39
MM1 pKa = 7.16MWTATLEE8 pKa = 4.01NSAIASRR15 pKa = 11.84QLQLSAVEE23 pKa = 3.95LWEE26 pKa = 4.98DD27 pKa = 3.41YY28 pKa = 11.17SYY30 pKa = 11.7LLNYY34 pKa = 9.55FFLLGVASVLAVVAIRR50 pKa = 11.84AAVNYY55 pKa = 10.07YY56 pKa = 10.61EE57 pKa = 4.31EE58 pKa = 4.98LRR60 pKa = 11.84LDD62 pKa = 3.61SLVRR66 pKa = 11.84EE67 pKa = 4.57LPTPPEE73 pKa = 4.25DD74 pKa = 3.53PVLDD78 pKa = 3.76GHH80 pKa = 7.62LEE82 pKa = 3.98FCPNNGYY89 pKa = 8.99VMRR92 pKa = 11.84VTTVIDD98 pKa = 4.0DD99 pKa = 3.99EE100 pKa = 4.42QHH102 pKa = 5.79SFLVKK107 pKa = 8.97VTPSQVTDD115 pKa = 3.47MLQGSSKK122 pKa = 8.06STRR125 pKa = 11.84VKK127 pKa = 10.05EE128 pKa = 4.07ARR130 pKa = 11.84GSSGVVPPGPRR141 pKa = 11.84AWDD144 pKa = 3.76AGNSTVPPAVHH155 pKa = 6.72LNGIPSAEE163 pKa = 3.87EE164 pKa = 3.43HH165 pKa = 7.18DD166 pKa = 3.66FDD168 pKa = 5.52LRR170 pKa = 11.84VVRR173 pKa = 11.84ALVRR177 pKa = 11.84MGVEE181 pKa = 3.7TRR183 pKa = 11.84EE184 pKa = 4.09NIQALGWDD192 pKa = 4.21FPEE195 pKa = 4.56ITASDD200 pKa = 4.03PQTPTTEE207 pKa = 4.04RR208 pKa = 11.84LLSEE212 pKa = 4.11TATSQEE218 pKa = 4.24LEE220 pKa = 4.11MAQPSSVVVPAQEE233 pKa = 4.46AFTKK237 pKa = 10.38KK238 pKa = 9.76ILKK241 pKa = 8.47ITDD244 pKa = 3.17RR245 pKa = 11.84EE246 pKa = 4.51GNVRR250 pKa = 11.84GLGFRR255 pKa = 11.84WEE257 pKa = 4.37NYY259 pKa = 9.72LVTAYY264 pKa = 10.05HH265 pKa = 6.34VYY267 pKa = 10.58EE268 pKa = 4.11SVKK271 pKa = 10.9NEE273 pKa = 3.92EE274 pKa = 4.73CDD276 pKa = 3.63VSPVPSTQGFPLSQFSALTYY296 pKa = 10.4SPKK299 pKa = 10.58LDD301 pKa = 4.29FIVLKK306 pKa = 10.37VDD308 pKa = 3.12KK309 pKa = 10.83PEE311 pKa = 4.08MVWAKK316 pKa = 10.39IGVSKK321 pKa = 10.64MSLKK325 pKa = 9.83TPAPNTPFKK334 pKa = 11.33VIGKK338 pKa = 8.9QNNSWNSSTGFLRR351 pKa = 11.84AAHH354 pKa = 6.79GDD356 pKa = 3.58LKK358 pKa = 10.57IKK360 pKa = 10.78HH361 pKa = 6.2NATTTKK367 pKa = 10.1GYY369 pKa = 10.2SGSPLLSDD377 pKa = 3.66GKK379 pKa = 11.15VVGLHH384 pKa = 6.62LGAANPTNYY393 pKa = 10.37CLDD396 pKa = 3.73LSFLPLLFQEE406 pKa = 5.05GVEE409 pKa = 4.24SEE411 pKa = 4.38EE412 pKa = 4.01KK413 pKa = 9.51QGRR416 pKa = 11.84YY417 pKa = 9.21HH418 pKa = 6.85KK419 pKa = 10.59FVDD422 pKa = 3.2SRR424 pKa = 11.84EE425 pKa = 4.27YY426 pKa = 10.33QDD428 pKa = 4.36HH429 pKa = 7.44ADD431 pKa = 5.2FKK433 pKa = 11.35VEE435 pKa = 3.41WEE437 pKa = 4.01EE438 pKa = 3.77RR439 pKa = 11.84RR440 pKa = 11.84KK441 pKa = 10.42EE442 pKa = 4.01EE443 pKa = 4.11RR444 pKa = 11.84DD445 pKa = 2.97EE446 pKa = 4.88DD447 pKa = 3.75YY448 pKa = 10.96TYY450 pKa = 11.85GLVHH454 pKa = 6.83NKK456 pKa = 9.69RR457 pKa = 11.84FIEE460 pKa = 3.65MRR462 pKa = 11.84YY463 pKa = 9.43RR464 pKa = 11.84KK465 pKa = 10.01SFGDD469 pKa = 3.42ATIVNASEE477 pKa = 3.69EE478 pKa = 4.24TTRR481 pKa = 11.84NFFNNKK487 pKa = 9.04ALPAGVRR494 pKa = 11.84WADD497 pKa = 3.28EE498 pKa = 4.07DD499 pKa = 4.66DD500 pKa = 4.49YY501 pKa = 11.46EE502 pKa = 5.75AVMPDD507 pKa = 3.94FVHH510 pKa = 7.56ASKK513 pKa = 10.78KK514 pKa = 7.96PQLRR518 pKa = 11.84GTEE521 pKa = 4.44TTCGMTQKK529 pKa = 10.8SSGPPPLVTSAPASVSSSSEE549 pKa = 3.75KK550 pKa = 10.62AKK552 pKa = 10.6PSAKK556 pKa = 8.48PTSPPTSILKK566 pKa = 9.61PSPKK570 pKa = 10.01LSSNDD575 pKa = 3.37QSSKK579 pKa = 9.48PSVVFKK585 pKa = 10.5EE586 pKa = 4.13VEE588 pKa = 4.02MQKK591 pKa = 10.56NSPSTSKK598 pKa = 10.01PASSGQPKK606 pKa = 9.73SRR608 pKa = 11.84KK609 pKa = 8.62SKK611 pKa = 10.56KK612 pKa = 9.58KK613 pKa = 8.3EE614 pKa = 3.45HH615 pKa = 6.14SQAFSAIIQNLEE627 pKa = 4.0SLSDD631 pKa = 3.74SEE633 pKa = 5.11VRR635 pKa = 11.84DD636 pKa = 3.46LKK638 pKa = 11.34SRR640 pKa = 11.84IGG642 pKa = 3.39
Molecular weight: 71.16 kDa
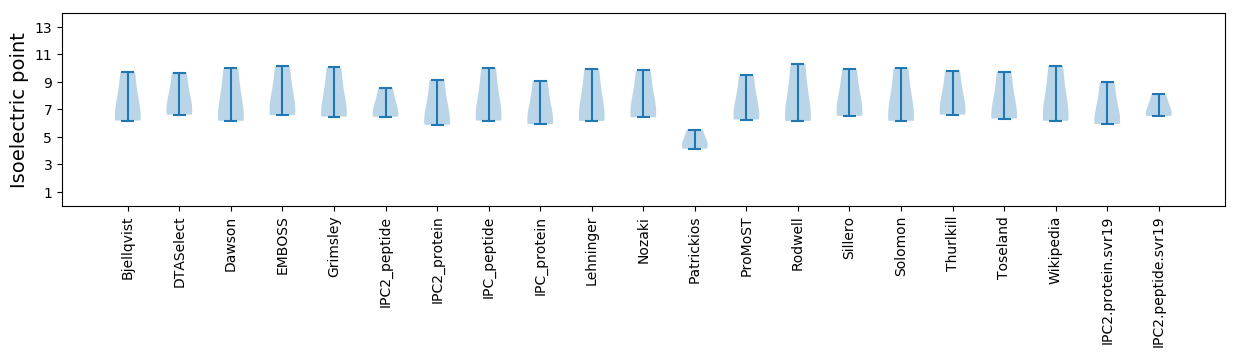
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KEZ9|A0A1L3KEZ9_9VIRU Peptidase S39 domain-containing protein OS=Hubei sobemo-like virus 2 OX=1923205 PE=4 SV=1
MM1 pKa = 7.54LNHH4 pKa = 7.55ASRR7 pKa = 11.84NLRR10 pKa = 11.84NSPMSTDD17 pKa = 3.2TFRR20 pKa = 11.84TSKK23 pKa = 10.52KK24 pKa = 9.22RR25 pKa = 11.84WPLFAPCLGGAGKK38 pKa = 10.63RR39 pKa = 11.84MPTKK43 pKa = 10.52RR44 pKa = 11.84KK45 pKa = 7.62QTSKK49 pKa = 10.43KK50 pKa = 10.12AKK52 pKa = 8.16STAKK56 pKa = 10.09PRR58 pKa = 11.84VQQRR62 pKa = 11.84QLAISQRR69 pKa = 11.84RR70 pKa = 11.84ARR72 pKa = 11.84PDD74 pKa = 2.96VGHH77 pKa = 6.83IVHH80 pKa = 6.74KK81 pKa = 10.91VCALTDD87 pKa = 4.1PFCPAAVGAKK97 pKa = 8.62WPNSTAIRR105 pKa = 11.84TLAYY109 pKa = 8.63PFHH112 pKa = 7.0SRR114 pKa = 11.84RR115 pKa = 11.84TVAVTATGAGAVIFTPGYY133 pKa = 9.79NNWTAGATIDD143 pKa = 4.97AEE145 pKa = 4.4TGNALFTNYY154 pKa = 8.07ATAPKK159 pKa = 10.23DD160 pKa = 2.87IGAALWRR167 pKa = 11.84IVSWGFKK174 pKa = 8.89IRR176 pKa = 11.84SVASPMLASGMVRR189 pKa = 11.84VRR191 pKa = 11.84IFAPAYY197 pKa = 10.2SGANNIDD204 pKa = 3.46PTSYY208 pKa = 10.53LCEE211 pKa = 4.34DD212 pKa = 5.36FIDD215 pKa = 5.91VPLQDD220 pKa = 4.42CKK222 pKa = 10.37EE223 pKa = 4.13LCVIPQRR230 pKa = 11.84SGEE233 pKa = 3.9LAEE236 pKa = 4.75IFNASDD242 pKa = 3.13VGGATIQSTANVGWPYY258 pKa = 11.41VLVSVDD264 pKa = 3.63GAPASTVPLDD274 pKa = 3.03IEE276 pKa = 4.4VYY278 pKa = 10.31YY279 pKa = 10.76NYY281 pKa = 10.67EE282 pKa = 4.22LMFSDD287 pKa = 5.07GNALSMLMTPSPSASGLMTQAASYY311 pKa = 7.37VTKK314 pKa = 10.78SLGSFVKK321 pKa = 10.39QGAKK325 pKa = 9.1QVEE328 pKa = 4.82GAVKK332 pKa = 8.8TAASRR337 pKa = 11.84YY338 pKa = 9.33IGGLLGGPMGAAAGGMLAITVDD360 pKa = 3.34
MM1 pKa = 7.54LNHH4 pKa = 7.55ASRR7 pKa = 11.84NLRR10 pKa = 11.84NSPMSTDD17 pKa = 3.2TFRR20 pKa = 11.84TSKK23 pKa = 10.52KK24 pKa = 9.22RR25 pKa = 11.84WPLFAPCLGGAGKK38 pKa = 10.63RR39 pKa = 11.84MPTKK43 pKa = 10.52RR44 pKa = 11.84KK45 pKa = 7.62QTSKK49 pKa = 10.43KK50 pKa = 10.12AKK52 pKa = 8.16STAKK56 pKa = 10.09PRR58 pKa = 11.84VQQRR62 pKa = 11.84QLAISQRR69 pKa = 11.84RR70 pKa = 11.84ARR72 pKa = 11.84PDD74 pKa = 2.96VGHH77 pKa = 6.83IVHH80 pKa = 6.74KK81 pKa = 10.91VCALTDD87 pKa = 4.1PFCPAAVGAKK97 pKa = 8.62WPNSTAIRR105 pKa = 11.84TLAYY109 pKa = 8.63PFHH112 pKa = 7.0SRR114 pKa = 11.84RR115 pKa = 11.84TVAVTATGAGAVIFTPGYY133 pKa = 9.79NNWTAGATIDD143 pKa = 4.97AEE145 pKa = 4.4TGNALFTNYY154 pKa = 8.07ATAPKK159 pKa = 10.23DD160 pKa = 2.87IGAALWRR167 pKa = 11.84IVSWGFKK174 pKa = 8.89IRR176 pKa = 11.84SVASPMLASGMVRR189 pKa = 11.84VRR191 pKa = 11.84IFAPAYY197 pKa = 10.2SGANNIDD204 pKa = 3.46PTSYY208 pKa = 10.53LCEE211 pKa = 4.34DD212 pKa = 5.36FIDD215 pKa = 5.91VPLQDD220 pKa = 4.42CKK222 pKa = 10.37EE223 pKa = 4.13LCVIPQRR230 pKa = 11.84SGEE233 pKa = 3.9LAEE236 pKa = 4.75IFNASDD242 pKa = 3.13VGGATIQSTANVGWPYY258 pKa = 11.41VLVSVDD264 pKa = 3.63GAPASTVPLDD274 pKa = 3.03IEE276 pKa = 4.4VYY278 pKa = 10.31YY279 pKa = 10.76NYY281 pKa = 10.67EE282 pKa = 4.22LMFSDD287 pKa = 5.07GNALSMLMTPSPSASGLMTQAASYY311 pKa = 7.37VTKK314 pKa = 10.78SLGSFVKK321 pKa = 10.39QGAKK325 pKa = 9.1QVEE328 pKa = 4.82GAVKK332 pKa = 8.8TAASRR337 pKa = 11.84YY338 pKa = 9.33IGGLLGGPMGAAAGGMLAITVDD360 pKa = 3.34
Molecular weight: 38.27 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1355 |
353 |
642 |
451.7 |
49.67 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.52 ± 1.639 | 1.328 ± 0.43 |
5.092 ± 0.581 | 5.609 ± 1.388 |
3.616 ± 0.106 | 6.199 ± 0.897 |
1.845 ± 0.278 | 3.838 ± 0.605 |
6.273 ± 0.482 | 8.118 ± 0.653 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.583 ± 0.423 | 4.207 ± 0.222 |
6.125 ± 0.691 | 3.469 ± 0.155 |
5.018 ± 0.199 | 9.299 ± 0.941 |
5.83 ± 0.928 | 7.306 ± 0.646 |
1.624 ± 0.146 | 3.1 ± 0.219 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
