
Kribbella amoyensis
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Propionibacteriales; Kribbellaceae; Kribbella
Average proteome isoelectric point is 6.22
Get precalculated fractions of proteins
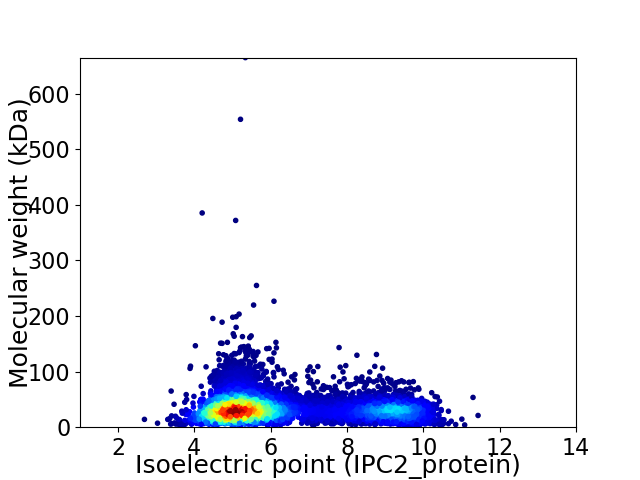
Virtual 2D-PAGE plot for 7393 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A561BT07|A0A561BT07_9ACTN Assimilatory nitrate reductase (NADH) alpha subunit apoprotein OS=Kribbella amoyensis OX=996641 GN=FB561_3154 PE=4 SV=1
MM1 pKa = 6.94FRR3 pKa = 11.84KK4 pKa = 9.75RR5 pKa = 11.84VAAAVMISGLAVATFGAINPASASEE30 pKa = 4.13SAPAAGSVRR39 pKa = 11.84MWEE42 pKa = 4.18DD43 pKa = 3.12PNYY46 pKa = 10.33TGSQYY51 pKa = 11.24VDD53 pKa = 3.36ATPANCPDD61 pKa = 3.45GCDD64 pKa = 3.23IDD66 pKa = 5.38GWDD69 pKa = 4.09GDD71 pKa = 4.15NEE73 pKa = 4.04ISSVRR78 pKa = 11.84NEE80 pKa = 4.09TTCTVRR86 pKa = 11.84LYY88 pKa = 11.21ADD90 pKa = 3.87DD91 pKa = 4.25NFGGRR96 pKa = 11.84SVDD99 pKa = 3.63LAAGEE104 pKa = 4.67SYY106 pKa = 11.63SNLEE110 pKa = 4.31LVGFDD115 pKa = 3.59NDD117 pKa = 3.59AEE119 pKa = 4.41SFQFVCC125 pKa = 5.56
MM1 pKa = 6.94FRR3 pKa = 11.84KK4 pKa = 9.75RR5 pKa = 11.84VAAAVMISGLAVATFGAINPASASEE30 pKa = 4.13SAPAAGSVRR39 pKa = 11.84MWEE42 pKa = 4.18DD43 pKa = 3.12PNYY46 pKa = 10.33TGSQYY51 pKa = 11.24VDD53 pKa = 3.36ATPANCPDD61 pKa = 3.45GCDD64 pKa = 3.23IDD66 pKa = 5.38GWDD69 pKa = 4.09GDD71 pKa = 4.15NEE73 pKa = 4.04ISSVRR78 pKa = 11.84NEE80 pKa = 4.09TTCTVRR86 pKa = 11.84LYY88 pKa = 11.21ADD90 pKa = 3.87DD91 pKa = 4.25NFGGRR96 pKa = 11.84SVDD99 pKa = 3.63LAAGEE104 pKa = 4.67SYY106 pKa = 11.63SNLEE110 pKa = 4.31LVGFDD115 pKa = 3.59NDD117 pKa = 3.59AEE119 pKa = 4.41SFQFVCC125 pKa = 5.56
Molecular weight: 13.23 kDa
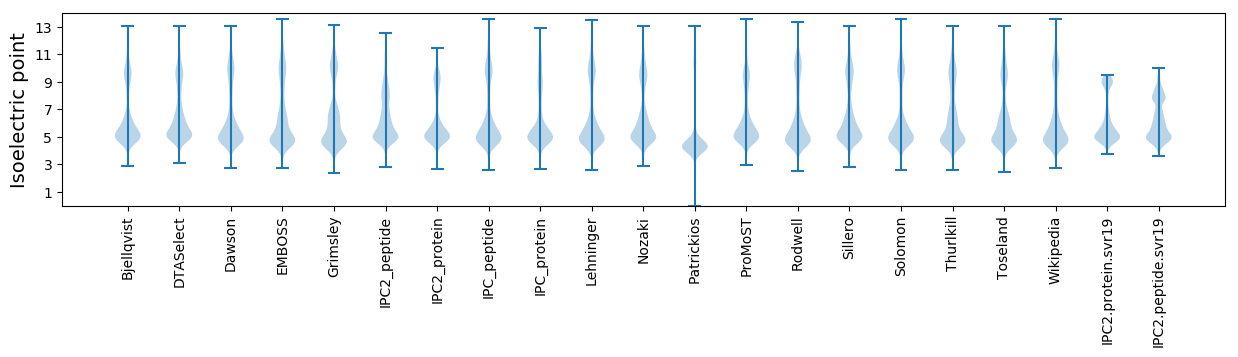
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A561BS86|A0A561BS86_9ACTN Nucleotide-binding universal stress UspA family protein OS=Kribbella amoyensis OX=996641 GN=FB561_2882 PE=3 SV=1
MM1 pKa = 7.51ARR3 pKa = 11.84PHH5 pKa = 5.96RR6 pKa = 11.84AGRR9 pKa = 11.84ASRR12 pKa = 11.84AVGGPSPPTGRR23 pKa = 11.84RR24 pKa = 11.84STCAPNRR31 pKa = 11.84PRR33 pKa = 11.84PARR36 pKa = 11.84APPRR40 pKa = 11.84TSRR43 pKa = 11.84PGTSRR48 pKa = 11.84PHH50 pKa = 6.21PRR52 pKa = 11.84RR53 pKa = 11.84NTSPVARR60 pKa = 11.84AQPVRR65 pKa = 11.84RR66 pKa = 11.84ARR68 pKa = 11.84RR69 pKa = 11.84VRR71 pKa = 11.84VVRR74 pKa = 11.84LVRR77 pKa = 11.84VRR79 pKa = 11.84RR80 pKa = 11.84VGSPRR85 pKa = 11.84GSPARR90 pKa = 11.84RR91 pKa = 11.84TTAPPVRR98 pKa = 11.84RR99 pKa = 11.84ARR101 pKa = 11.84ARR103 pKa = 11.84ARR105 pKa = 11.84ARR107 pKa = 11.84ARR109 pKa = 11.84ARR111 pKa = 11.84ARR113 pKa = 11.84ARR115 pKa = 11.84ARR117 pKa = 11.84APQVSRR123 pKa = 11.84APRR126 pKa = 11.84PRR128 pKa = 11.84QVPAGRR134 pKa = 11.84VRR136 pKa = 11.84QVPPATTLPPAPPARR151 pKa = 11.84ADD153 pKa = 3.48RR154 pKa = 11.84PTRR157 pKa = 11.84VRR159 pKa = 11.84VAPALLGRR167 pKa = 11.84MALGLLGRR175 pKa = 11.84AVRR178 pKa = 11.84LGLLRR183 pKa = 11.84ARR185 pKa = 11.84LRR187 pKa = 11.84RR188 pKa = 11.84ARR190 pKa = 11.84GTSRR194 pKa = 11.84CPGRR198 pKa = 11.84AGPTVRR204 pKa = 11.84AVSTLPRR211 pKa = 11.84RR212 pKa = 11.84RR213 pKa = 11.84ATSARR218 pKa = 11.84TRR220 pKa = 11.84ATTPPPPVGDD230 pKa = 3.53RR231 pKa = 11.84LRR233 pKa = 11.84ALVGAAPRR241 pKa = 11.84RR242 pKa = 11.84TPAGDD247 pKa = 3.47RR248 pKa = 11.84QQGRR252 pKa = 11.84ATTTRR257 pKa = 11.84PRR259 pKa = 11.84APAGVRR265 pKa = 11.84PRR267 pKa = 11.84ARR269 pKa = 11.84AATARR274 pKa = 11.84RR275 pKa = 11.84RR276 pKa = 11.84APVPVRR282 pKa = 11.84QAVRR286 pKa = 11.84VTAALGRR293 pKa = 11.84VPAGDD298 pKa = 3.97LLRR301 pKa = 11.84GVLRR305 pKa = 11.84RR306 pKa = 11.84MPAGVLPVALRR317 pKa = 11.84VVLRR321 pKa = 11.84RR322 pKa = 11.84ARR324 pKa = 11.84VGVPRR329 pKa = 11.84TVPRR333 pKa = 11.84RR334 pKa = 11.84VLGGVRR340 pKa = 11.84GPVVPGRR347 pKa = 11.84TRR349 pKa = 11.84AGGLLTVPVAGVPRR363 pKa = 11.84RR364 pKa = 11.84APGGVLPRR372 pKa = 11.84VRR374 pKa = 11.84ALGLVPPATGRR385 pKa = 11.84AARR388 pKa = 11.84PGRR391 pKa = 11.84SRR393 pKa = 11.84RR394 pKa = 11.84RR395 pKa = 11.84LRR397 pKa = 11.84APGPPAPGTKK407 pKa = 10.25DD408 pKa = 3.34PAKK411 pKa = 10.79DD412 pKa = 3.09PATRR416 pKa = 11.84AQAVRR421 pKa = 11.84APRR424 pKa = 11.84TKK426 pKa = 10.82APVTRR431 pKa = 11.84GPVTRR436 pKa = 11.84GRR438 pKa = 11.84RR439 pKa = 11.84VGRR442 pKa = 11.84ATRR445 pKa = 11.84GRR447 pKa = 11.84GPGPPGFRR455 pKa = 11.84LTSSRR460 pKa = 11.84VVGRR464 pKa = 11.84APASSLPGSSRR475 pKa = 11.84PGSTRR480 pKa = 11.84RR481 pKa = 11.84SRR483 pKa = 11.84FRR485 pKa = 11.84SRR487 pKa = 11.84RR488 pKa = 11.84VVAAAASSGRR498 pKa = 11.84RR499 pKa = 11.84RR500 pKa = 11.84RR501 pKa = 11.84ACC503 pKa = 3.64
MM1 pKa = 7.51ARR3 pKa = 11.84PHH5 pKa = 5.96RR6 pKa = 11.84AGRR9 pKa = 11.84ASRR12 pKa = 11.84AVGGPSPPTGRR23 pKa = 11.84RR24 pKa = 11.84STCAPNRR31 pKa = 11.84PRR33 pKa = 11.84PARR36 pKa = 11.84APPRR40 pKa = 11.84TSRR43 pKa = 11.84PGTSRR48 pKa = 11.84PHH50 pKa = 6.21PRR52 pKa = 11.84RR53 pKa = 11.84NTSPVARR60 pKa = 11.84AQPVRR65 pKa = 11.84RR66 pKa = 11.84ARR68 pKa = 11.84RR69 pKa = 11.84VRR71 pKa = 11.84VVRR74 pKa = 11.84LVRR77 pKa = 11.84VRR79 pKa = 11.84RR80 pKa = 11.84VGSPRR85 pKa = 11.84GSPARR90 pKa = 11.84RR91 pKa = 11.84TTAPPVRR98 pKa = 11.84RR99 pKa = 11.84ARR101 pKa = 11.84ARR103 pKa = 11.84ARR105 pKa = 11.84ARR107 pKa = 11.84ARR109 pKa = 11.84ARR111 pKa = 11.84ARR113 pKa = 11.84ARR115 pKa = 11.84ARR117 pKa = 11.84APQVSRR123 pKa = 11.84APRR126 pKa = 11.84PRR128 pKa = 11.84QVPAGRR134 pKa = 11.84VRR136 pKa = 11.84QVPPATTLPPAPPARR151 pKa = 11.84ADD153 pKa = 3.48RR154 pKa = 11.84PTRR157 pKa = 11.84VRR159 pKa = 11.84VAPALLGRR167 pKa = 11.84MALGLLGRR175 pKa = 11.84AVRR178 pKa = 11.84LGLLRR183 pKa = 11.84ARR185 pKa = 11.84LRR187 pKa = 11.84RR188 pKa = 11.84ARR190 pKa = 11.84GTSRR194 pKa = 11.84CPGRR198 pKa = 11.84AGPTVRR204 pKa = 11.84AVSTLPRR211 pKa = 11.84RR212 pKa = 11.84RR213 pKa = 11.84ATSARR218 pKa = 11.84TRR220 pKa = 11.84ATTPPPPVGDD230 pKa = 3.53RR231 pKa = 11.84LRR233 pKa = 11.84ALVGAAPRR241 pKa = 11.84RR242 pKa = 11.84TPAGDD247 pKa = 3.47RR248 pKa = 11.84QQGRR252 pKa = 11.84ATTTRR257 pKa = 11.84PRR259 pKa = 11.84APAGVRR265 pKa = 11.84PRR267 pKa = 11.84ARR269 pKa = 11.84AATARR274 pKa = 11.84RR275 pKa = 11.84RR276 pKa = 11.84APVPVRR282 pKa = 11.84QAVRR286 pKa = 11.84VTAALGRR293 pKa = 11.84VPAGDD298 pKa = 3.97LLRR301 pKa = 11.84GVLRR305 pKa = 11.84RR306 pKa = 11.84MPAGVLPVALRR317 pKa = 11.84VVLRR321 pKa = 11.84RR322 pKa = 11.84ARR324 pKa = 11.84VGVPRR329 pKa = 11.84TVPRR333 pKa = 11.84RR334 pKa = 11.84VLGGVRR340 pKa = 11.84GPVVPGRR347 pKa = 11.84TRR349 pKa = 11.84AGGLLTVPVAGVPRR363 pKa = 11.84RR364 pKa = 11.84APGGVLPRR372 pKa = 11.84VRR374 pKa = 11.84ALGLVPPATGRR385 pKa = 11.84AARR388 pKa = 11.84PGRR391 pKa = 11.84SRR393 pKa = 11.84RR394 pKa = 11.84RR395 pKa = 11.84LRR397 pKa = 11.84APGPPAPGTKK407 pKa = 10.25DD408 pKa = 3.34PAKK411 pKa = 10.79DD412 pKa = 3.09PATRR416 pKa = 11.84AQAVRR421 pKa = 11.84APRR424 pKa = 11.84TKK426 pKa = 10.82APVTRR431 pKa = 11.84GPVTRR436 pKa = 11.84GRR438 pKa = 11.84RR439 pKa = 11.84VGRR442 pKa = 11.84ATRR445 pKa = 11.84GRR447 pKa = 11.84GPGPPGFRR455 pKa = 11.84LTSSRR460 pKa = 11.84VVGRR464 pKa = 11.84APASSLPGSSRR475 pKa = 11.84PGSTRR480 pKa = 11.84RR481 pKa = 11.84SRR483 pKa = 11.84FRR485 pKa = 11.84SRR487 pKa = 11.84RR488 pKa = 11.84VVAAAASSGRR498 pKa = 11.84RR499 pKa = 11.84RR500 pKa = 11.84RR501 pKa = 11.84ACC503 pKa = 3.64
Molecular weight: 53.59 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2439600 |
27 |
6241 |
330.0 |
35.46 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.85 ± 0.033 | 0.679 ± 0.008 |
6.132 ± 0.027 | 5.463 ± 0.032 |
2.895 ± 0.017 | 9.079 ± 0.027 |
2.032 ± 0.013 | 3.656 ± 0.019 |
2.299 ± 0.022 | 10.529 ± 0.041 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.625 ± 0.009 | 2.025 ± 0.018 |
5.822 ± 0.024 | 3.086 ± 0.017 |
7.464 ± 0.033 | 5.23 ± 0.022 |
6.397 ± 0.028 | 8.854 ± 0.026 |
1.641 ± 0.012 | 2.241 ± 0.014 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
