
Halorhabdus tiamatea SARL4B
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Stenosarchaea group; Halobacteria; Halobacteriales; Haloarculaceae; Halorhabdus; Halorhabdus tiamatea
Average proteome isoelectric point is 4.84
Get precalculated fractions of proteins
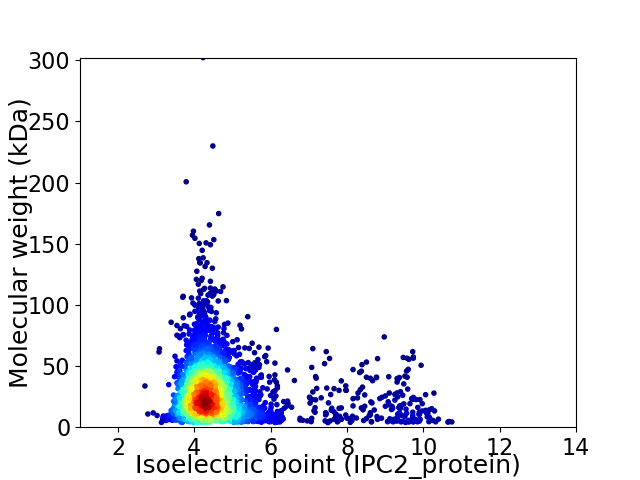
Virtual 2D-PAGE plot for 2988 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|F7PJY1|F7PJY1_9EURY Uncharacterized protein OS=Halorhabdus tiamatea SARL4B OX=1033806 GN=HLRTI_000228 PE=4 SV=1
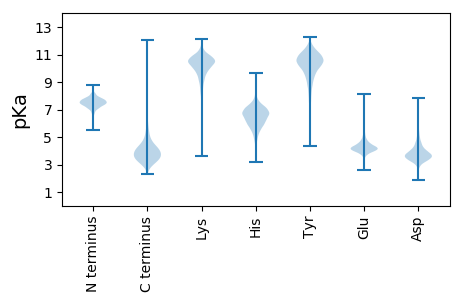
MM1 pKa = 7.19TAGDD5 pKa = 4.0DD6 pKa = 3.53THH8 pKa = 6.28TLNRR12 pKa = 11.84RR13 pKa = 11.84HH14 pKa = 5.16VLKK17 pKa = 10.86SIGAGAVGTTSVLSARR33 pKa = 11.84SVIADD38 pKa = 3.32SDD40 pKa = 3.75GTHH43 pKa = 6.31YY44 pKa = 11.26SNPLYY49 pKa = 10.87GPDD52 pKa = 3.56FADD55 pKa = 3.7PTIHH59 pKa = 7.44RR60 pKa = 11.84ADD62 pKa = 4.54DD63 pKa = 4.08GTWWAYY69 pKa = 9.48ASNMSYY75 pKa = 10.41IDD77 pKa = 4.11DD78 pKa = 4.2ADD80 pKa = 4.01EE81 pKa = 4.12EE82 pKa = 4.55LVPILSSPDD91 pKa = 3.43LVNWTYY97 pKa = 11.0EE98 pKa = 3.92GEE100 pKa = 4.36AFDD103 pKa = 6.02SRR105 pKa = 11.84PGWLYY110 pKa = 11.14GSIWAPDD117 pKa = 3.32VHH119 pKa = 7.3YY120 pKa = 10.98HH121 pKa = 6.79DD122 pKa = 4.99GQWVLFYY129 pKa = 11.1ALWPRR134 pKa = 11.84GDD136 pKa = 5.28DD137 pKa = 3.63DD138 pKa = 4.37SQVPGIGVATADD150 pKa = 3.91TPDD153 pKa = 4.16GPFTDD158 pKa = 3.71HH159 pKa = 7.54GEE161 pKa = 4.06ILSNPDD167 pKa = 2.78HH168 pKa = 7.49PYY170 pKa = 10.15PGNTIDD176 pKa = 4.34PYY178 pKa = 11.19FVFHH182 pKa = 7.16NEE184 pKa = 3.48TPYY187 pKa = 11.26LFWANFAGINVVEE200 pKa = 4.37LTDD203 pKa = 3.99DD204 pKa = 3.61LRR206 pKa = 11.84DD207 pKa = 3.71YY208 pKa = 11.09RR209 pKa = 11.84AGTFDD214 pKa = 3.75QIAGSAYY221 pKa = 9.49EE222 pKa = 4.37GPAIFKK228 pKa = 10.68RR229 pKa = 11.84GDD231 pKa = 2.96YY232 pKa = 9.67WYY234 pKa = 10.91VFGSTGDD241 pKa = 3.62CCDD244 pKa = 4.4GFDD247 pKa = 3.29STYY250 pKa = 9.61EE251 pKa = 4.04VEE253 pKa = 4.46VGRR256 pKa = 11.84SEE258 pKa = 4.74YY259 pKa = 10.75FLGPYY264 pKa = 9.23HH265 pKa = 7.15DD266 pKa = 5.05RR267 pKa = 11.84NGTPMLEE274 pKa = 3.76RR275 pKa = 11.84DD276 pKa = 3.57EE277 pKa = 4.74WNAGPTHH284 pKa = 6.96LGDD287 pKa = 3.34NDD289 pKa = 3.74RR290 pKa = 11.84FVGPGHH296 pKa = 7.07GDD298 pKa = 3.24VTVDD302 pKa = 4.74DD303 pKa = 5.63DD304 pKa = 3.69GTYY307 pKa = 9.13WFVYY311 pKa = 9.18HH312 pKa = 7.5AYY314 pKa = 8.92DD315 pKa = 3.46TEE317 pKa = 4.96GPAFVDD323 pKa = 3.95SGWPPARR330 pKa = 11.84QLFVDD335 pKa = 4.87PIRR338 pKa = 11.84WEE340 pKa = 4.44DD341 pKa = 3.08GWPIIGCDD349 pKa = 3.6GTPSIEE355 pKa = 4.04MPIPGEE361 pKa = 4.0GGYY364 pKa = 10.81CSGSDD369 pKa = 4.44DD370 pKa = 3.74GTGDD374 pKa = 3.37GGGDD378 pKa = 3.61GTDD381 pKa = 3.76DD382 pKa = 5.26RR383 pKa = 11.84YY384 pKa = 9.71EE385 pKa = 4.15TWSDD389 pKa = 2.96TDD391 pKa = 3.51DD392 pKa = 4.27PYY394 pKa = 11.93YY395 pKa = 10.11STLVSDD401 pKa = 4.23LEE403 pKa = 4.59GYY405 pKa = 10.15NLASAGQFVNGNEE418 pKa = 4.16EE419 pKa = 3.85ATVWDD424 pKa = 4.21TYY426 pKa = 11.24EE427 pKa = 4.34IDD429 pKa = 3.73GGNADD434 pKa = 3.89RR435 pKa = 11.84LEE437 pKa = 4.28RR438 pKa = 11.84SSLAVSDD445 pKa = 4.14EE446 pKa = 4.52VPFSEE451 pKa = 4.23AARR454 pKa = 11.84FEE456 pKa = 4.5VTDD459 pKa = 3.63TPEE462 pKa = 4.24NPWEE466 pKa = 4.03ITLKK470 pKa = 10.79SVGEE474 pKa = 4.09RR475 pKa = 11.84EE476 pKa = 4.3LEE478 pKa = 4.21SGHH481 pKa = 6.12VLLGVAYY488 pKa = 9.27MRR490 pKa = 11.84TPDD493 pKa = 3.42EE494 pKa = 4.38EE495 pKa = 5.31ASVTYY500 pKa = 10.55KK501 pKa = 9.81STASSNEE508 pKa = 3.2SDD510 pKa = 3.6NYY512 pKa = 7.88VTRR515 pKa = 11.84AQPPLDD521 pKa = 4.17TEE523 pKa = 3.81WQRR526 pKa = 11.84YY527 pKa = 5.32YY528 pKa = 11.03FPIEE532 pKa = 4.32FGVSAAPGEE541 pKa = 4.22WWTEE545 pKa = 3.29IWLGAAAQTVDD556 pKa = 2.83IGGLAVIDD564 pKa = 4.11FARR567 pKa = 11.84GVRR570 pKa = 11.84ADD572 pKa = 4.26DD573 pKa = 4.22LPVGEE578 pKa = 5.46DD579 pKa = 3.24PGEE582 pKa = 4.25TAVEE586 pKa = 4.1TDD588 pKa = 4.28DD589 pKa = 3.66GTEE592 pKa = 3.97TVTSGEE598 pKa = 4.42SEE600 pKa = 4.31GEE602 pKa = 3.8QDD604 pKa = 5.46DD605 pKa = 5.76DD606 pKa = 3.92EE607 pKa = 4.98TSDD610 pKa = 4.88SEE612 pKa = 4.42GTPADD617 pKa = 4.11ALPGGEE623 pKa = 5.05GSSQDD628 pKa = 3.42LDD630 pKa = 3.78GDD632 pKa = 4.07GLYY635 pKa = 10.83EE636 pKa = 5.18DD637 pKa = 4.61VDD639 pKa = 4.15GDD641 pKa = 3.75GDD643 pKa = 4.34ADD645 pKa = 3.49ISDD648 pKa = 3.75VRR650 pKa = 11.84SLLTNFNSEE659 pKa = 4.33MVQEE663 pKa = 4.57HH664 pKa = 6.85ADD666 pKa = 3.51AYY668 pKa = 11.04DD669 pKa = 3.35FDD671 pKa = 6.51GDD673 pKa = 3.93GGVDD677 pKa = 3.4VGDD680 pKa = 3.54VLALYY685 pKa = 9.77RR686 pKa = 11.84RR687 pKa = 11.84SYY689 pKa = 11.13RR690 pKa = 11.84SVTT693 pKa = 3.29
MM1 pKa = 7.19TAGDD5 pKa = 4.0DD6 pKa = 3.53THH8 pKa = 6.28TLNRR12 pKa = 11.84RR13 pKa = 11.84HH14 pKa = 5.16VLKK17 pKa = 10.86SIGAGAVGTTSVLSARR33 pKa = 11.84SVIADD38 pKa = 3.32SDD40 pKa = 3.75GTHH43 pKa = 6.31YY44 pKa = 11.26SNPLYY49 pKa = 10.87GPDD52 pKa = 3.56FADD55 pKa = 3.7PTIHH59 pKa = 7.44RR60 pKa = 11.84ADD62 pKa = 4.54DD63 pKa = 4.08GTWWAYY69 pKa = 9.48ASNMSYY75 pKa = 10.41IDD77 pKa = 4.11DD78 pKa = 4.2ADD80 pKa = 4.01EE81 pKa = 4.12EE82 pKa = 4.55LVPILSSPDD91 pKa = 3.43LVNWTYY97 pKa = 11.0EE98 pKa = 3.92GEE100 pKa = 4.36AFDD103 pKa = 6.02SRR105 pKa = 11.84PGWLYY110 pKa = 11.14GSIWAPDD117 pKa = 3.32VHH119 pKa = 7.3YY120 pKa = 10.98HH121 pKa = 6.79DD122 pKa = 4.99GQWVLFYY129 pKa = 11.1ALWPRR134 pKa = 11.84GDD136 pKa = 5.28DD137 pKa = 3.63DD138 pKa = 4.37SQVPGIGVATADD150 pKa = 3.91TPDD153 pKa = 4.16GPFTDD158 pKa = 3.71HH159 pKa = 7.54GEE161 pKa = 4.06ILSNPDD167 pKa = 2.78HH168 pKa = 7.49PYY170 pKa = 10.15PGNTIDD176 pKa = 4.34PYY178 pKa = 11.19FVFHH182 pKa = 7.16NEE184 pKa = 3.48TPYY187 pKa = 11.26LFWANFAGINVVEE200 pKa = 4.37LTDD203 pKa = 3.99DD204 pKa = 3.61LRR206 pKa = 11.84DD207 pKa = 3.71YY208 pKa = 11.09RR209 pKa = 11.84AGTFDD214 pKa = 3.75QIAGSAYY221 pKa = 9.49EE222 pKa = 4.37GPAIFKK228 pKa = 10.68RR229 pKa = 11.84GDD231 pKa = 2.96YY232 pKa = 9.67WYY234 pKa = 10.91VFGSTGDD241 pKa = 3.62CCDD244 pKa = 4.4GFDD247 pKa = 3.29STYY250 pKa = 9.61EE251 pKa = 4.04VEE253 pKa = 4.46VGRR256 pKa = 11.84SEE258 pKa = 4.74YY259 pKa = 10.75FLGPYY264 pKa = 9.23HH265 pKa = 7.15DD266 pKa = 5.05RR267 pKa = 11.84NGTPMLEE274 pKa = 3.76RR275 pKa = 11.84DD276 pKa = 3.57EE277 pKa = 4.74WNAGPTHH284 pKa = 6.96LGDD287 pKa = 3.34NDD289 pKa = 3.74RR290 pKa = 11.84FVGPGHH296 pKa = 7.07GDD298 pKa = 3.24VTVDD302 pKa = 4.74DD303 pKa = 5.63DD304 pKa = 3.69GTYY307 pKa = 9.13WFVYY311 pKa = 9.18HH312 pKa = 7.5AYY314 pKa = 8.92DD315 pKa = 3.46TEE317 pKa = 4.96GPAFVDD323 pKa = 3.95SGWPPARR330 pKa = 11.84QLFVDD335 pKa = 4.87PIRR338 pKa = 11.84WEE340 pKa = 4.44DD341 pKa = 3.08GWPIIGCDD349 pKa = 3.6GTPSIEE355 pKa = 4.04MPIPGEE361 pKa = 4.0GGYY364 pKa = 10.81CSGSDD369 pKa = 4.44DD370 pKa = 3.74GTGDD374 pKa = 3.37GGGDD378 pKa = 3.61GTDD381 pKa = 3.76DD382 pKa = 5.26RR383 pKa = 11.84YY384 pKa = 9.71EE385 pKa = 4.15TWSDD389 pKa = 2.96TDD391 pKa = 3.51DD392 pKa = 4.27PYY394 pKa = 11.93YY395 pKa = 10.11STLVSDD401 pKa = 4.23LEE403 pKa = 4.59GYY405 pKa = 10.15NLASAGQFVNGNEE418 pKa = 4.16EE419 pKa = 3.85ATVWDD424 pKa = 4.21TYY426 pKa = 11.24EE427 pKa = 4.34IDD429 pKa = 3.73GGNADD434 pKa = 3.89RR435 pKa = 11.84LEE437 pKa = 4.28RR438 pKa = 11.84SSLAVSDD445 pKa = 4.14EE446 pKa = 4.52VPFSEE451 pKa = 4.23AARR454 pKa = 11.84FEE456 pKa = 4.5VTDD459 pKa = 3.63TPEE462 pKa = 4.24NPWEE466 pKa = 4.03ITLKK470 pKa = 10.79SVGEE474 pKa = 4.09RR475 pKa = 11.84EE476 pKa = 4.3LEE478 pKa = 4.21SGHH481 pKa = 6.12VLLGVAYY488 pKa = 9.27MRR490 pKa = 11.84TPDD493 pKa = 3.42EE494 pKa = 4.38EE495 pKa = 5.31ASVTYY500 pKa = 10.55KK501 pKa = 9.81STASSNEE508 pKa = 3.2SDD510 pKa = 3.6NYY512 pKa = 7.88VTRR515 pKa = 11.84AQPPLDD521 pKa = 4.17TEE523 pKa = 3.81WQRR526 pKa = 11.84YY527 pKa = 5.32YY528 pKa = 11.03FPIEE532 pKa = 4.32FGVSAAPGEE541 pKa = 4.22WWTEE545 pKa = 3.29IWLGAAAQTVDD556 pKa = 2.83IGGLAVIDD564 pKa = 4.11FARR567 pKa = 11.84GVRR570 pKa = 11.84ADD572 pKa = 4.26DD573 pKa = 4.22LPVGEE578 pKa = 5.46DD579 pKa = 3.24PGEE582 pKa = 4.25TAVEE586 pKa = 4.1TDD588 pKa = 4.28DD589 pKa = 3.66GTEE592 pKa = 3.97TVTSGEE598 pKa = 4.42SEE600 pKa = 4.31GEE602 pKa = 3.8QDD604 pKa = 5.46DD605 pKa = 5.76DD606 pKa = 3.92EE607 pKa = 4.98TSDD610 pKa = 4.88SEE612 pKa = 4.42GTPADD617 pKa = 4.11ALPGGEE623 pKa = 5.05GSSQDD628 pKa = 3.42LDD630 pKa = 3.78GDD632 pKa = 4.07GLYY635 pKa = 10.83EE636 pKa = 5.18DD637 pKa = 4.61VDD639 pKa = 4.15GDD641 pKa = 3.75GDD643 pKa = 4.34ADD645 pKa = 3.49ISDD648 pKa = 3.75VRR650 pKa = 11.84SLLTNFNSEE659 pKa = 4.33MVQEE663 pKa = 4.57HH664 pKa = 6.85ADD666 pKa = 3.51AYY668 pKa = 11.04DD669 pKa = 3.35FDD671 pKa = 6.51GDD673 pKa = 3.93GGVDD677 pKa = 3.4VGDD680 pKa = 3.54VLALYY685 pKa = 9.77RR686 pKa = 11.84RR687 pKa = 11.84SYY689 pKa = 11.13RR690 pKa = 11.84SVTT693 pKa = 3.29
Molecular weight: 75.68 kDa
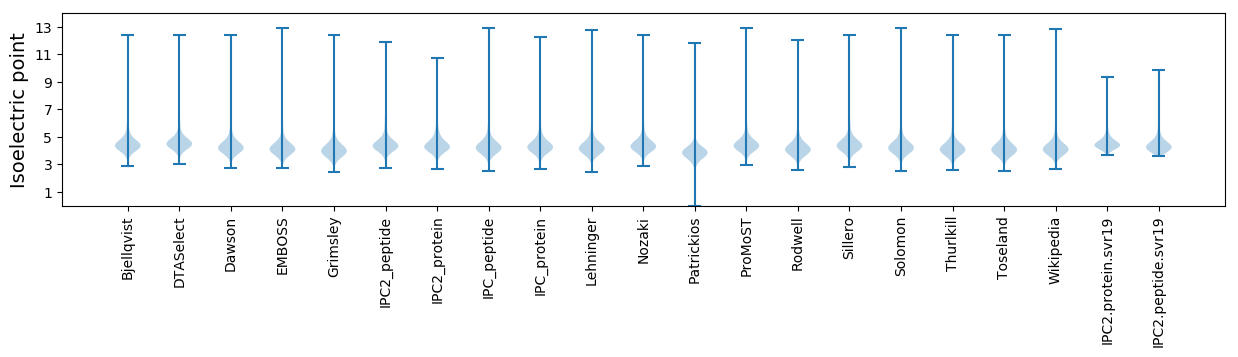
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|F7PNT0|F7PNT0_9EURY Pyruvate synthase subunit PorA protein OS=Halorhabdus tiamatea SARL4B OX=1033806 GN=porA PE=4 SV=1
MM1 pKa = 7.13ATSSEE6 pKa = 3.88KK7 pKa = 10.42RR8 pKa = 11.84EE9 pKa = 3.73RR10 pKa = 11.84AMAAMVKK17 pKa = 9.99HH18 pKa = 5.38YY19 pKa = 10.57GEE21 pKa = 5.0RR22 pKa = 11.84RR23 pKa = 11.84QQLNPFDD30 pKa = 4.38EE31 pKa = 4.43GRR33 pKa = 11.84KK34 pKa = 8.47YY35 pKa = 10.54HH36 pKa = 7.35DD37 pKa = 4.13SFAVGYY43 pKa = 9.52SGGYY47 pKa = 8.65AASFLVPVAAEE58 pKa = 3.94AKK60 pKa = 9.63AAQAAARR67 pKa = 11.84MPYY70 pKa = 8.85MAALLRR76 pKa = 11.84SVNTMKK82 pKa = 10.7RR83 pKa = 11.84SIKK86 pKa = 10.45GVTIGSAMWTGGKK99 pKa = 8.87IARR102 pKa = 11.84RR103 pKa = 11.84MPDD106 pKa = 2.24VDD108 pKa = 4.22FNVGRR113 pKa = 11.84RR114 pKa = 11.84VSDD117 pKa = 3.54VEE119 pKa = 3.67LDD121 pKa = 3.47GRR123 pKa = 11.84AKK125 pKa = 10.63QIVGQTADD133 pKa = 3.57TLRR136 pKa = 11.84NTPLPRR142 pKa = 11.84QRR144 pKa = 11.84RR145 pKa = 11.84IAKK148 pKa = 7.69LTRR151 pKa = 11.84NTGDD155 pKa = 3.4GDD157 pKa = 3.73VMEE160 pKa = 4.43WLTRR164 pKa = 11.84NVEE167 pKa = 4.15APDD170 pKa = 3.32SEE172 pKa = 4.77RR173 pKa = 11.84YY174 pKa = 8.36QRR176 pKa = 11.84GLVYY180 pKa = 10.7LRR182 pKa = 11.84RR183 pKa = 11.84TGAEE187 pKa = 4.14GKK189 pKa = 9.74QLLSRR194 pKa = 11.84LTQDD198 pKa = 3.94ARR200 pKa = 11.84SALLKK205 pKa = 10.9LRR207 pKa = 11.84DD208 pKa = 3.49KK209 pKa = 11.43AGLQRR214 pKa = 11.84QLTKK218 pKa = 10.47AWVRR222 pKa = 11.84GDD224 pKa = 3.42IDD226 pKa = 4.04ASDD229 pKa = 3.16IGKK232 pKa = 8.64ATRR235 pKa = 11.84RR236 pKa = 11.84YY237 pKa = 10.42DD238 pKa = 3.46GLDD241 pKa = 3.21AEE243 pKa = 4.99GRR245 pKa = 11.84AAFDD249 pKa = 4.92DD250 pKa = 4.41LVQHH254 pKa = 6.77EE255 pKa = 5.12GPDD258 pKa = 3.67AVSVSGKK265 pKa = 9.92LDD267 pKa = 3.23EE268 pKa = 5.71DD269 pKa = 3.7AFEE272 pKa = 5.46DD273 pKa = 3.96VLKK276 pKa = 10.98SDD278 pKa = 4.58LSTARR283 pKa = 11.84KK284 pKa = 9.71ADD286 pKa = 3.39IVDD289 pKa = 3.76SLSEE293 pKa = 3.97VDD295 pKa = 4.61QMQSFNTSTIEE306 pKa = 4.01LVSRR310 pKa = 11.84PSGWRR315 pKa = 11.84GTCAGIVWRR324 pKa = 11.84RR325 pKa = 11.84SFAGPTPTRR334 pKa = 11.84RR335 pKa = 11.84TSWPTGRR342 pKa = 11.84ANWMCSSTVPSPMRR356 pKa = 11.84SRR358 pKa = 11.84TVTLAVMSSRR368 pKa = 11.84RR369 pKa = 11.84RR370 pKa = 11.84STRR373 pKa = 11.84SMSWRR378 pKa = 11.84TSGGRR383 pKa = 11.84VRR385 pKa = 11.84ANSASVSSSNHH396 pKa = 4.16PTAAVSSSPPSTTPRR411 pKa = 11.84FTNSVPKK418 pKa = 10.66
MM1 pKa = 7.13ATSSEE6 pKa = 3.88KK7 pKa = 10.42RR8 pKa = 11.84EE9 pKa = 3.73RR10 pKa = 11.84AMAAMVKK17 pKa = 9.99HH18 pKa = 5.38YY19 pKa = 10.57GEE21 pKa = 5.0RR22 pKa = 11.84RR23 pKa = 11.84QQLNPFDD30 pKa = 4.38EE31 pKa = 4.43GRR33 pKa = 11.84KK34 pKa = 8.47YY35 pKa = 10.54HH36 pKa = 7.35DD37 pKa = 4.13SFAVGYY43 pKa = 9.52SGGYY47 pKa = 8.65AASFLVPVAAEE58 pKa = 3.94AKK60 pKa = 9.63AAQAAARR67 pKa = 11.84MPYY70 pKa = 8.85MAALLRR76 pKa = 11.84SVNTMKK82 pKa = 10.7RR83 pKa = 11.84SIKK86 pKa = 10.45GVTIGSAMWTGGKK99 pKa = 8.87IARR102 pKa = 11.84RR103 pKa = 11.84MPDD106 pKa = 2.24VDD108 pKa = 4.22FNVGRR113 pKa = 11.84RR114 pKa = 11.84VSDD117 pKa = 3.54VEE119 pKa = 3.67LDD121 pKa = 3.47GRR123 pKa = 11.84AKK125 pKa = 10.63QIVGQTADD133 pKa = 3.57TLRR136 pKa = 11.84NTPLPRR142 pKa = 11.84QRR144 pKa = 11.84RR145 pKa = 11.84IAKK148 pKa = 7.69LTRR151 pKa = 11.84NTGDD155 pKa = 3.4GDD157 pKa = 3.73VMEE160 pKa = 4.43WLTRR164 pKa = 11.84NVEE167 pKa = 4.15APDD170 pKa = 3.32SEE172 pKa = 4.77RR173 pKa = 11.84YY174 pKa = 8.36QRR176 pKa = 11.84GLVYY180 pKa = 10.7LRR182 pKa = 11.84RR183 pKa = 11.84TGAEE187 pKa = 4.14GKK189 pKa = 9.74QLLSRR194 pKa = 11.84LTQDD198 pKa = 3.94ARR200 pKa = 11.84SALLKK205 pKa = 10.9LRR207 pKa = 11.84DD208 pKa = 3.49KK209 pKa = 11.43AGLQRR214 pKa = 11.84QLTKK218 pKa = 10.47AWVRR222 pKa = 11.84GDD224 pKa = 3.42IDD226 pKa = 4.04ASDD229 pKa = 3.16IGKK232 pKa = 8.64ATRR235 pKa = 11.84RR236 pKa = 11.84YY237 pKa = 10.42DD238 pKa = 3.46GLDD241 pKa = 3.21AEE243 pKa = 4.99GRR245 pKa = 11.84AAFDD249 pKa = 4.92DD250 pKa = 4.41LVQHH254 pKa = 6.77EE255 pKa = 5.12GPDD258 pKa = 3.67AVSVSGKK265 pKa = 9.92LDD267 pKa = 3.23EE268 pKa = 5.71DD269 pKa = 3.7AFEE272 pKa = 5.46DD273 pKa = 3.96VLKK276 pKa = 10.98SDD278 pKa = 4.58LSTARR283 pKa = 11.84KK284 pKa = 9.71ADD286 pKa = 3.39IVDD289 pKa = 3.76SLSEE293 pKa = 3.97VDD295 pKa = 4.61QMQSFNTSTIEE306 pKa = 4.01LVSRR310 pKa = 11.84PSGWRR315 pKa = 11.84GTCAGIVWRR324 pKa = 11.84RR325 pKa = 11.84SFAGPTPTRR334 pKa = 11.84RR335 pKa = 11.84TSWPTGRR342 pKa = 11.84ANWMCSSTVPSPMRR356 pKa = 11.84SRR358 pKa = 11.84TVTLAVMSSRR368 pKa = 11.84RR369 pKa = 11.84RR370 pKa = 11.84STRR373 pKa = 11.84SMSWRR378 pKa = 11.84TSGGRR383 pKa = 11.84VRR385 pKa = 11.84ANSASVSSSNHH396 pKa = 4.16PTAAVSSSPPSTTPRR411 pKa = 11.84FTNSVPKK418 pKa = 10.66
Molecular weight: 45.94 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
856988 |
37 |
2728 |
286.8 |
31.32 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.501 ± 0.062 | 0.639 ± 0.017 |
8.697 ± 0.047 | 8.956 ± 0.063 |
3.269 ± 0.027 | 8.201 ± 0.05 |
2.049 ± 0.025 | 4.393 ± 0.031 |
1.857 ± 0.03 | 8.802 ± 0.059 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.737 ± 0.02 | 2.399 ± 0.032 |
4.592 ± 0.029 | 2.729 ± 0.027 |
6.467 ± 0.047 | 5.493 ± 0.033 |
6.566 ± 0.043 | 8.709 ± 0.047 |
1.186 ± 0.02 | 2.759 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
