
Capybara microvirus Cap3_SP_541
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 5.33
Get precalculated fractions of proteins
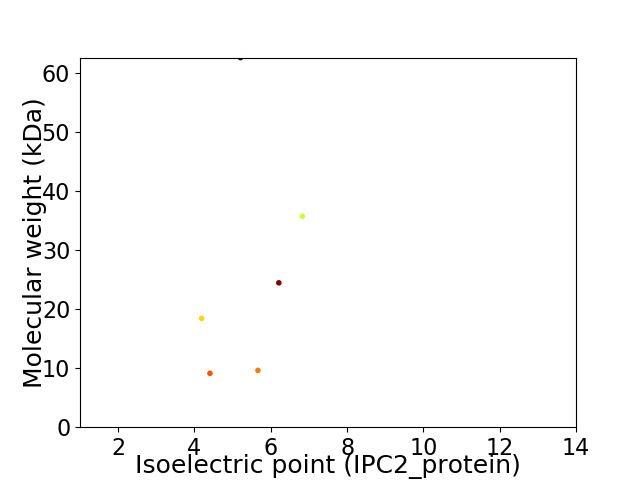
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4P8W5D5|A0A4P8W5D5_9VIRU Major capsid protein OS=Capybara microvirus Cap3_SP_541 OX=2585472 PE=3 SV=1
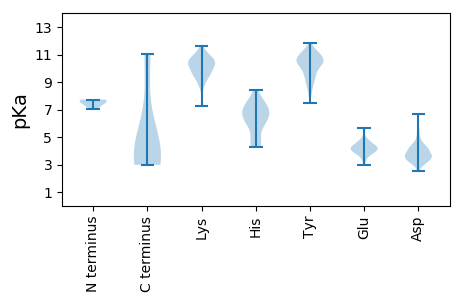
MM1 pKa = 7.73SLEE4 pKa = 4.02SALIPLRR11 pKa = 11.84LLSGHH16 pKa = 5.98SANTSFSDD24 pKa = 3.2PGSPIKK30 pKa = 10.33QLYY33 pKa = 8.35HH34 pKa = 6.75GKK36 pKa = 10.27YY37 pKa = 9.8DD38 pKa = 3.86DD39 pKa = 3.92NGRR42 pKa = 11.84LTLVPDD48 pKa = 4.09GEE50 pKa = 4.78INLQEE55 pKa = 4.41QIDD58 pKa = 4.17SFRR61 pKa = 11.84DD62 pKa = 3.44QCDD65 pKa = 2.51MDD67 pKa = 4.24YY68 pKa = 10.6IIRR71 pKa = 11.84QLLQGNEE78 pKa = 4.25SVLSSRR84 pKa = 11.84QSLGYY89 pKa = 9.75IDD91 pKa = 3.52TTEE94 pKa = 4.46YY95 pKa = 9.2PSSMAALLQLRR106 pKa = 11.84IDD108 pKa = 4.13AEE110 pKa = 4.72SIFYY114 pKa = 10.43SLPLDD119 pKa = 3.86EE120 pKa = 5.44RR121 pKa = 11.84QHH123 pKa = 6.48FDD125 pKa = 2.69NSFDD129 pKa = 3.24VWFAQTGSPDD139 pKa = 3.14WLKK142 pKa = 11.04SISSSVSSSDD152 pKa = 3.15PAAPAAPAEE161 pKa = 4.49VISNEE166 pKa = 4.12SS167 pKa = 2.98
MM1 pKa = 7.73SLEE4 pKa = 4.02SALIPLRR11 pKa = 11.84LLSGHH16 pKa = 5.98SANTSFSDD24 pKa = 3.2PGSPIKK30 pKa = 10.33QLYY33 pKa = 8.35HH34 pKa = 6.75GKK36 pKa = 10.27YY37 pKa = 9.8DD38 pKa = 3.86DD39 pKa = 3.92NGRR42 pKa = 11.84LTLVPDD48 pKa = 4.09GEE50 pKa = 4.78INLQEE55 pKa = 4.41QIDD58 pKa = 4.17SFRR61 pKa = 11.84DD62 pKa = 3.44QCDD65 pKa = 2.51MDD67 pKa = 4.24YY68 pKa = 10.6IIRR71 pKa = 11.84QLLQGNEE78 pKa = 4.25SVLSSRR84 pKa = 11.84QSLGYY89 pKa = 9.75IDD91 pKa = 3.52TTEE94 pKa = 4.46YY95 pKa = 9.2PSSMAALLQLRR106 pKa = 11.84IDD108 pKa = 4.13AEE110 pKa = 4.72SIFYY114 pKa = 10.43SLPLDD119 pKa = 3.86EE120 pKa = 5.44RR121 pKa = 11.84QHH123 pKa = 6.48FDD125 pKa = 2.69NSFDD129 pKa = 3.24VWFAQTGSPDD139 pKa = 3.14WLKK142 pKa = 11.04SISSSVSSSDD152 pKa = 3.15PAAPAAPAEE161 pKa = 4.49VISNEE166 pKa = 4.12SS167 pKa = 2.98
Molecular weight: 18.45 kDa
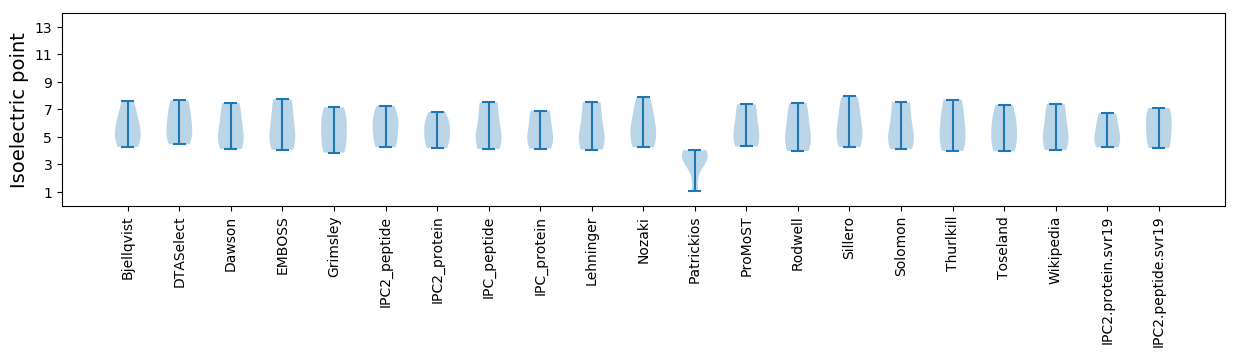
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4V1FVU5|A0A4V1FVU5_9VIRU Replication initiator protein OS=Capybara microvirus Cap3_SP_541 OX=2585472 PE=4 SV=1
MM1 pKa = 7.42SCYY4 pKa = 10.36SPNVVRR10 pKa = 11.84WSADD14 pKa = 3.01PFTGEE19 pKa = 4.72ASAQFLGQLRR29 pKa = 11.84DD30 pKa = 3.53QYY32 pKa = 11.24TYY34 pKa = 11.87DD35 pKa = 3.51HH36 pKa = 6.99LEE38 pKa = 4.0EE39 pKa = 3.99QAMQLVPCGQCIGCRR54 pKa = 11.84LDD56 pKa = 4.34YY57 pKa = 10.66SRR59 pKa = 11.84HH60 pKa = 4.39WADD63 pKa = 3.54RR64 pKa = 11.84LVLEE68 pKa = 4.31LSKK71 pKa = 11.59YY72 pKa = 10.07NGSALFITLTYY83 pKa = 10.92NDD85 pKa = 4.13DD86 pKa = 4.25HH87 pKa = 7.6LPLSDD92 pKa = 4.49DD93 pKa = 3.92FSVPTLLVRR102 pKa = 11.84DD103 pKa = 3.64TQLYY107 pKa = 9.7HH108 pKa = 6.32KK109 pKa = 9.54RR110 pKa = 11.84LRR112 pKa = 11.84KK113 pKa = 9.7AFPDD117 pKa = 3.03RR118 pKa = 11.84VLRR121 pKa = 11.84FFMCGEE127 pKa = 4.23YY128 pKa = 10.98GSITHH133 pKa = 6.84RR134 pKa = 11.84PHH136 pKa = 5.93YY137 pKa = 10.61HH138 pKa = 5.78EE139 pKa = 4.64VLFGLSMCDD148 pKa = 4.06FSDD151 pKa = 4.91LRR153 pKa = 11.84PCQRR157 pKa = 11.84NEE159 pKa = 3.57LGQLSFTSPTLEE171 pKa = 4.71KK172 pKa = 9.7IWSNGFILFSEE183 pKa = 4.36ISYY186 pKa = 11.46ASMCYY191 pKa = 9.54VSRR194 pKa = 11.84YY195 pKa = 7.49SLKK198 pKa = 10.72KK199 pKa = 10.45AFGYY203 pKa = 10.97SNDD206 pKa = 3.61PRR208 pKa = 11.84QHH210 pKa = 6.7PEE212 pKa = 3.48FLLMSRR218 pKa = 11.84RR219 pKa = 11.84PGIGLDD225 pKa = 3.43YY226 pKa = 10.91FEE228 pKa = 5.65SHH230 pKa = 7.49DD231 pKa = 3.67PTDD234 pKa = 3.43VVFFSGKK241 pKa = 9.81KK242 pKa = 8.66ISIPRR247 pKa = 11.84SVLDD251 pKa = 3.44RR252 pKa = 11.84CGDD255 pKa = 3.55SAFVSKK261 pKa = 10.84VLEE264 pKa = 3.89QRR266 pKa = 11.84QRR268 pKa = 11.84LAHH271 pKa = 6.38DD272 pKa = 3.56TLLTEE277 pKa = 5.68LSLTDD282 pKa = 5.32LPYY285 pKa = 10.86LDD287 pKa = 4.24YY288 pKa = 11.14IKK290 pKa = 11.02VKK292 pKa = 10.1EE293 pKa = 3.84RR294 pKa = 11.84SKK296 pKa = 11.25AEE298 pKa = 4.18KK299 pKa = 9.61IAKK302 pKa = 9.25LKK304 pKa = 10.54KK305 pKa = 10.4RR306 pKa = 11.84SDD308 pKa = 3.41LL309 pKa = 3.78
MM1 pKa = 7.42SCYY4 pKa = 10.36SPNVVRR10 pKa = 11.84WSADD14 pKa = 3.01PFTGEE19 pKa = 4.72ASAQFLGQLRR29 pKa = 11.84DD30 pKa = 3.53QYY32 pKa = 11.24TYY34 pKa = 11.87DD35 pKa = 3.51HH36 pKa = 6.99LEE38 pKa = 4.0EE39 pKa = 3.99QAMQLVPCGQCIGCRR54 pKa = 11.84LDD56 pKa = 4.34YY57 pKa = 10.66SRR59 pKa = 11.84HH60 pKa = 4.39WADD63 pKa = 3.54RR64 pKa = 11.84LVLEE68 pKa = 4.31LSKK71 pKa = 11.59YY72 pKa = 10.07NGSALFITLTYY83 pKa = 10.92NDD85 pKa = 4.13DD86 pKa = 4.25HH87 pKa = 7.6LPLSDD92 pKa = 4.49DD93 pKa = 3.92FSVPTLLVRR102 pKa = 11.84DD103 pKa = 3.64TQLYY107 pKa = 9.7HH108 pKa = 6.32KK109 pKa = 9.54RR110 pKa = 11.84LRR112 pKa = 11.84KK113 pKa = 9.7AFPDD117 pKa = 3.03RR118 pKa = 11.84VLRR121 pKa = 11.84FFMCGEE127 pKa = 4.23YY128 pKa = 10.98GSITHH133 pKa = 6.84RR134 pKa = 11.84PHH136 pKa = 5.93YY137 pKa = 10.61HH138 pKa = 5.78EE139 pKa = 4.64VLFGLSMCDD148 pKa = 4.06FSDD151 pKa = 4.91LRR153 pKa = 11.84PCQRR157 pKa = 11.84NEE159 pKa = 3.57LGQLSFTSPTLEE171 pKa = 4.71KK172 pKa = 9.7IWSNGFILFSEE183 pKa = 4.36ISYY186 pKa = 11.46ASMCYY191 pKa = 9.54VSRR194 pKa = 11.84YY195 pKa = 7.49SLKK198 pKa = 10.72KK199 pKa = 10.45AFGYY203 pKa = 10.97SNDD206 pKa = 3.61PRR208 pKa = 11.84QHH210 pKa = 6.7PEE212 pKa = 3.48FLLMSRR218 pKa = 11.84RR219 pKa = 11.84PGIGLDD225 pKa = 3.43YY226 pKa = 10.91FEE228 pKa = 5.65SHH230 pKa = 7.49DD231 pKa = 3.67PTDD234 pKa = 3.43VVFFSGKK241 pKa = 9.81KK242 pKa = 8.66ISIPRR247 pKa = 11.84SVLDD251 pKa = 3.44RR252 pKa = 11.84CGDD255 pKa = 3.55SAFVSKK261 pKa = 10.84VLEE264 pKa = 3.89QRR266 pKa = 11.84QRR268 pKa = 11.84LAHH271 pKa = 6.38DD272 pKa = 3.56TLLTEE277 pKa = 5.68LSLTDD282 pKa = 5.32LPYY285 pKa = 10.86LDD287 pKa = 4.24YY288 pKa = 11.14IKK290 pKa = 11.02VKK292 pKa = 10.1EE293 pKa = 3.84RR294 pKa = 11.84SKK296 pKa = 11.25AEE298 pKa = 4.18KK299 pKa = 9.61IAKK302 pKa = 9.25LKK304 pKa = 10.54KK305 pKa = 10.4RR306 pKa = 11.84SDD308 pKa = 3.41LL309 pKa = 3.78
Molecular weight: 35.77 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1437 |
77 |
556 |
239.5 |
26.68 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.933 ± 2.048 | 1.253 ± 0.494 |
6.959 ± 0.803 | 3.967 ± 0.45 |
5.428 ± 0.733 | 6.054 ± 0.809 |
1.949 ± 0.437 | 4.384 ± 0.507 |
3.619 ± 0.472 | 8.838 ± 1.452 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.088 ± 0.394 | 3.688 ± 0.613 |
5.498 ± 0.416 | 4.036 ± 0.449 |
5.08 ± 0.865 | 12.178 ± 1.551 |
5.08 ± 0.774 | 5.289 ± 0.527 |
1.253 ± 0.204 | 5.428 ± 0.564 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
