
Kosmotoga olearia (strain ATCC BAA-1733 / DSM 21960 / TBF 19.5.1)
Taxonomy: cellular organisms; Bacteria; Thermotogae; Thermotogae; Kosmotogales; Kosmotogaceae; Kosmotoga; Kosmotoga olearia
Average proteome isoelectric point is 6.8
Get precalculated fractions of proteins
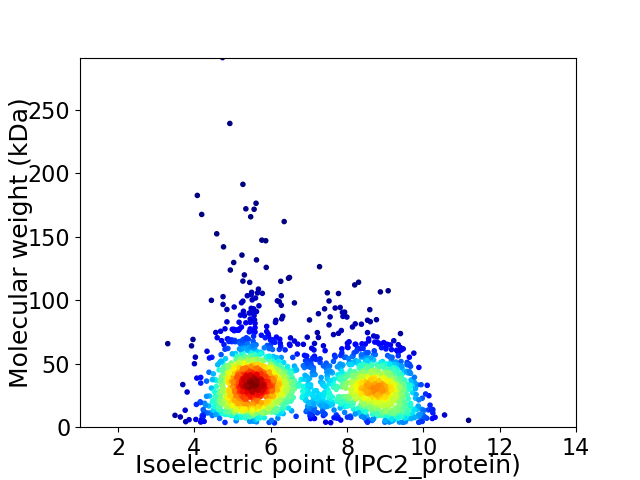
Virtual 2D-PAGE plot for 2087 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|C5CI24|C5CI24_KOSOT Uncharacterized protein OS=Kosmotoga olearia (strain ATCC BAA-1733 / DSM 21960 / TBF 19.5.1) OX=521045 GN=Kole_1101 PE=4 SV=1
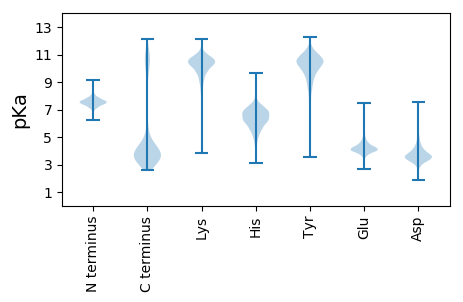
MM1 pKa = 7.6KK2 pKa = 10.67NFFGPVLLFLVLLFASCHH20 pKa = 6.27PIDD23 pKa = 4.91NPIDD27 pKa = 4.05GDD29 pKa = 4.09GLILFDD35 pKa = 4.61EE36 pKa = 5.06GHH38 pKa = 6.53AQTAGNADD46 pKa = 3.33WVIDD50 pKa = 3.67GGYY53 pKa = 9.94SEE55 pKa = 5.66FADD58 pKa = 3.38ALKK61 pKa = 10.75EE62 pKa = 3.79KK63 pKa = 9.94GYY65 pKa = 10.59SVEE68 pKa = 4.26STDD71 pKa = 5.64DD72 pKa = 3.75PFDD75 pKa = 3.97YY76 pKa = 10.12STLARR81 pKa = 11.84YY82 pKa = 9.41DD83 pKa = 3.41VVIIPEE89 pKa = 3.97PNIRR93 pKa = 11.84FSSDD97 pKa = 3.07EE98 pKa = 3.53ISALKK103 pKa = 10.25RR104 pKa = 11.84YY105 pKa = 9.24VEE107 pKa = 4.23NGGSVFFIGNHH118 pKa = 5.93SGADD122 pKa = 3.65RR123 pKa = 11.84NNDD126 pKa = 2.38GWDD129 pKa = 3.24PVEE132 pKa = 4.39IFNEE136 pKa = 4.23VVNDD140 pKa = 3.71DD141 pKa = 3.43FGFRR145 pKa = 11.84FDD147 pKa = 4.48SNTVSEE153 pKa = 5.34DD154 pKa = 3.84PIEE157 pKa = 5.49DD158 pKa = 3.38ILVTPITYY166 pKa = 9.95GVNAVGMWAGSTITIINDD184 pKa = 3.52SKK186 pKa = 10.05VHH188 pKa = 4.59VAIRR192 pKa = 11.84AYY194 pKa = 8.54EE195 pKa = 3.99KK196 pKa = 10.1PYY198 pKa = 11.02VVYY201 pKa = 9.99GYY203 pKa = 9.92YY204 pKa = 10.91GSGKK208 pKa = 9.57FVAIGDD214 pKa = 4.02SSPFDD219 pKa = 5.35DD220 pKa = 5.62GDD222 pKa = 3.96GDD224 pKa = 4.43PGDD227 pKa = 3.87NLYY230 pKa = 11.21DD231 pKa = 3.14NWYY234 pKa = 10.22DD235 pKa = 3.66YY236 pKa = 11.52DD237 pKa = 4.92DD238 pKa = 4.34SVLAVNIVDD247 pKa = 3.95WLSKK251 pKa = 10.84
MM1 pKa = 7.6KK2 pKa = 10.67NFFGPVLLFLVLLFASCHH20 pKa = 6.27PIDD23 pKa = 4.91NPIDD27 pKa = 4.05GDD29 pKa = 4.09GLILFDD35 pKa = 4.61EE36 pKa = 5.06GHH38 pKa = 6.53AQTAGNADD46 pKa = 3.33WVIDD50 pKa = 3.67GGYY53 pKa = 9.94SEE55 pKa = 5.66FADD58 pKa = 3.38ALKK61 pKa = 10.75EE62 pKa = 3.79KK63 pKa = 9.94GYY65 pKa = 10.59SVEE68 pKa = 4.26STDD71 pKa = 5.64DD72 pKa = 3.75PFDD75 pKa = 3.97YY76 pKa = 10.12STLARR81 pKa = 11.84YY82 pKa = 9.41DD83 pKa = 3.41VVIIPEE89 pKa = 3.97PNIRR93 pKa = 11.84FSSDD97 pKa = 3.07EE98 pKa = 3.53ISALKK103 pKa = 10.25RR104 pKa = 11.84YY105 pKa = 9.24VEE107 pKa = 4.23NGGSVFFIGNHH118 pKa = 5.93SGADD122 pKa = 3.65RR123 pKa = 11.84NNDD126 pKa = 2.38GWDD129 pKa = 3.24PVEE132 pKa = 4.39IFNEE136 pKa = 4.23VVNDD140 pKa = 3.71DD141 pKa = 3.43FGFRR145 pKa = 11.84FDD147 pKa = 4.48SNTVSEE153 pKa = 5.34DD154 pKa = 3.84PIEE157 pKa = 5.49DD158 pKa = 3.38ILVTPITYY166 pKa = 9.95GVNAVGMWAGSTITIINDD184 pKa = 3.52SKK186 pKa = 10.05VHH188 pKa = 4.59VAIRR192 pKa = 11.84AYY194 pKa = 8.54EE195 pKa = 3.99KK196 pKa = 10.1PYY198 pKa = 11.02VVYY201 pKa = 9.99GYY203 pKa = 9.92YY204 pKa = 10.91GSGKK208 pKa = 9.57FVAIGDD214 pKa = 4.02SSPFDD219 pKa = 5.35DD220 pKa = 5.62GDD222 pKa = 3.96GDD224 pKa = 4.43PGDD227 pKa = 3.87NLYY230 pKa = 11.21DD231 pKa = 3.14NWYY234 pKa = 10.22DD235 pKa = 3.66YY236 pKa = 11.52DD237 pKa = 4.92DD238 pKa = 4.34SVLAVNIVDD247 pKa = 3.95WLSKK251 pKa = 10.84
Molecular weight: 27.73 kDa
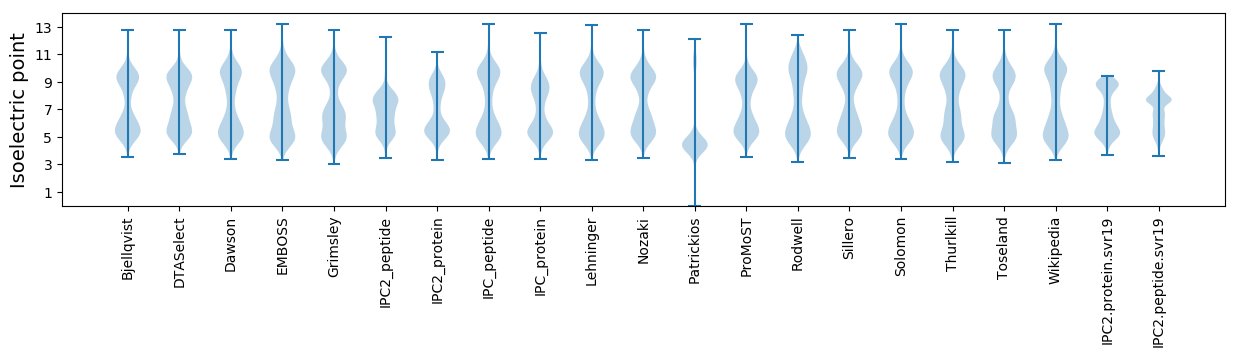
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|C5CD82|C5CD82_KOSOT Binding-protein-dependent transport systems inner membrane component OS=Kosmotoga olearia (strain ATCC BAA-1733 / DSM 21960 / TBF 19.5.1) OX=521045 GN=Kole_0301 PE=3 SV=1
MM1 pKa = 7.35KK2 pKa = 9.4RR3 pKa = 11.84TYY5 pKa = 10.16QPSRR9 pKa = 11.84VKK11 pKa = 10.48RR12 pKa = 11.84KK13 pKa = 7.72RR14 pKa = 11.84THH16 pKa = 5.76GFLVRR21 pKa = 11.84MRR23 pKa = 11.84TKK25 pKa = 9.87SGRR28 pKa = 11.84RR29 pKa = 11.84IIANRR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 8.89GRR39 pKa = 11.84KK40 pKa = 8.69RR41 pKa = 11.84LAVV44 pKa = 3.41
MM1 pKa = 7.35KK2 pKa = 9.4RR3 pKa = 11.84TYY5 pKa = 10.16QPSRR9 pKa = 11.84VKK11 pKa = 10.48RR12 pKa = 11.84KK13 pKa = 7.72RR14 pKa = 11.84THH16 pKa = 5.76GFLVRR21 pKa = 11.84MRR23 pKa = 11.84TKK25 pKa = 9.87SGRR28 pKa = 11.84RR29 pKa = 11.84IIANRR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 8.89GRR39 pKa = 11.84KK40 pKa = 8.69RR41 pKa = 11.84LAVV44 pKa = 3.41
Molecular weight: 5.41 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
667979 |
31 |
2628 |
320.1 |
36.18 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.289 ± 0.048 | 0.749 ± 0.019 |
5.05 ± 0.043 | 7.85 ± 0.062 |
5.065 ± 0.044 | 6.849 ± 0.051 |
1.577 ± 0.019 | 8.258 ± 0.049 |
7.806 ± 0.051 | 10.125 ± 0.065 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.408 ± 0.025 | 4.13 ± 0.029 |
3.835 ± 0.03 | 2.147 ± 0.022 |
4.894 ± 0.036 | 6.006 ± 0.04 |
4.863 ± 0.035 | 7.285 ± 0.045 |
1.009 ± 0.02 | 3.804 ± 0.033 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
