
Candidatus Nasuia deltocephalinicola str. NAS-ALF
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Betaproteobacteria; Betaproteobacteria incertae sedis; Candidatus Nasuia; Candidatus Nasuia deltocephalinicola
Average proteome isoelectric point is 8.85
Get precalculated fractions of proteins
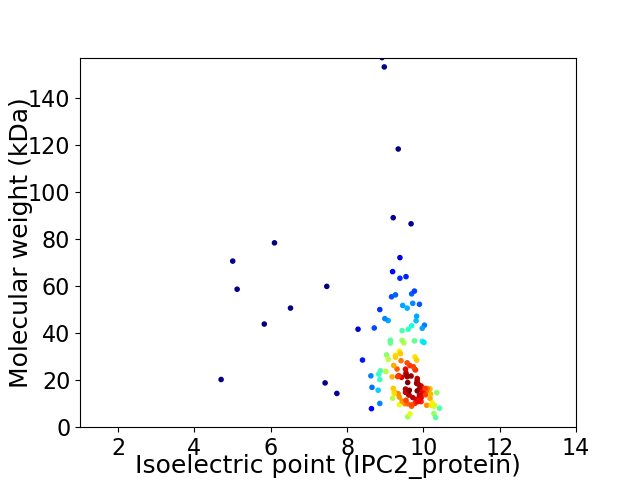
Virtual 2D-PAGE plot for 137 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|S5SQE3|S5SQE3_9PROT Cytochrome c oxidase subunit I OS=Candidatus Nasuia deltocephalinicola str. NAS-ALF OX=1343077 GN=cyoB PE=3 SV=1
MM1 pKa = 7.06FNKK4 pKa = 9.72VILMGEE10 pKa = 3.77IAMDD14 pKa = 3.6PEE16 pKa = 4.04LRR18 pKa = 11.84YY19 pKa = 10.2FKK21 pKa = 10.37DD22 pKa = 3.18NSVITVIKK30 pKa = 10.64LITRR34 pKa = 11.84EE35 pKa = 4.09NINNIKK41 pKa = 10.09DD42 pKa = 4.13LQWHH46 pKa = 6.68RR47 pKa = 11.84IILKK51 pKa = 9.59EE52 pKa = 3.94KK53 pKa = 10.99NKK55 pKa = 9.98ILKK58 pKa = 10.14KK59 pKa = 10.83NMIILIEE66 pKa = 4.2GKK68 pKa = 9.76IKK70 pKa = 9.01TRR72 pKa = 11.84RR73 pKa = 11.84WMDD76 pKa = 3.26NVKK79 pKa = 10.4YY80 pKa = 10.87NYY82 pKa = 8.26ITEE85 pKa = 4.27IFADD89 pKa = 4.04NIKK92 pKa = 10.51ILDD95 pKa = 3.64NKK97 pKa = 10.55NYY99 pKa = 10.68NNYY102 pKa = 9.64KK103 pKa = 9.99NKK105 pKa = 10.42KK106 pKa = 8.87NDD108 pKa = 3.69YY109 pKa = 9.85IDD111 pKa = 4.21NDD113 pKa = 3.59EE114 pKa = 5.02KK115 pKa = 11.54EE116 pKa = 4.58EE117 pKa = 4.12NDD119 pKa = 3.9EE120 pKa = 3.94NDD122 pKa = 3.32NYY124 pKa = 11.29NLEE127 pKa = 4.27DD128 pKa = 4.08SEE130 pKa = 4.83YY131 pKa = 10.27EE132 pKa = 4.14NKK134 pKa = 10.11NSKK137 pKa = 9.57IDD139 pKa = 3.73EE140 pKa = 5.19LIDD143 pKa = 4.0DD144 pKa = 4.65EE145 pKa = 5.33KK146 pKa = 11.83DD147 pKa = 3.38NIITVGGDD155 pKa = 3.53TEE157 pKa = 4.34LKK159 pKa = 9.98EE160 pKa = 3.92YY161 pKa = 10.71SYY163 pKa = 11.81DD164 pKa = 3.62EE165 pKa = 5.27DD166 pKa = 4.39YY167 pKa = 11.35EE168 pKa = 4.19
MM1 pKa = 7.06FNKK4 pKa = 9.72VILMGEE10 pKa = 3.77IAMDD14 pKa = 3.6PEE16 pKa = 4.04LRR18 pKa = 11.84YY19 pKa = 10.2FKK21 pKa = 10.37DD22 pKa = 3.18NSVITVIKK30 pKa = 10.64LITRR34 pKa = 11.84EE35 pKa = 4.09NINNIKK41 pKa = 10.09DD42 pKa = 4.13LQWHH46 pKa = 6.68RR47 pKa = 11.84IILKK51 pKa = 9.59EE52 pKa = 3.94KK53 pKa = 10.99NKK55 pKa = 9.98ILKK58 pKa = 10.14KK59 pKa = 10.83NMIILIEE66 pKa = 4.2GKK68 pKa = 9.76IKK70 pKa = 9.01TRR72 pKa = 11.84RR73 pKa = 11.84WMDD76 pKa = 3.26NVKK79 pKa = 10.4YY80 pKa = 10.87NYY82 pKa = 8.26ITEE85 pKa = 4.27IFADD89 pKa = 4.04NIKK92 pKa = 10.51ILDD95 pKa = 3.64NKK97 pKa = 10.55NYY99 pKa = 10.68NNYY102 pKa = 9.64KK103 pKa = 9.99NKK105 pKa = 10.42KK106 pKa = 8.87NDD108 pKa = 3.69YY109 pKa = 9.85IDD111 pKa = 4.21NDD113 pKa = 3.59EE114 pKa = 5.02KK115 pKa = 11.54EE116 pKa = 4.58EE117 pKa = 4.12NDD119 pKa = 3.9EE120 pKa = 3.94NDD122 pKa = 3.32NYY124 pKa = 11.29NLEE127 pKa = 4.27DD128 pKa = 4.08SEE130 pKa = 4.83YY131 pKa = 10.27EE132 pKa = 4.14NKK134 pKa = 10.11NSKK137 pKa = 9.57IDD139 pKa = 3.73EE140 pKa = 5.19LIDD143 pKa = 4.0DD144 pKa = 4.65EE145 pKa = 5.33KK146 pKa = 11.83DD147 pKa = 3.38NIITVGGDD155 pKa = 3.53TEE157 pKa = 4.34LKK159 pKa = 9.98EE160 pKa = 3.92YY161 pKa = 10.71SYY163 pKa = 11.81DD164 pKa = 3.62EE165 pKa = 5.27DD166 pKa = 4.39YY167 pKa = 11.35EE168 pKa = 4.19
Molecular weight: 20.29 kDa
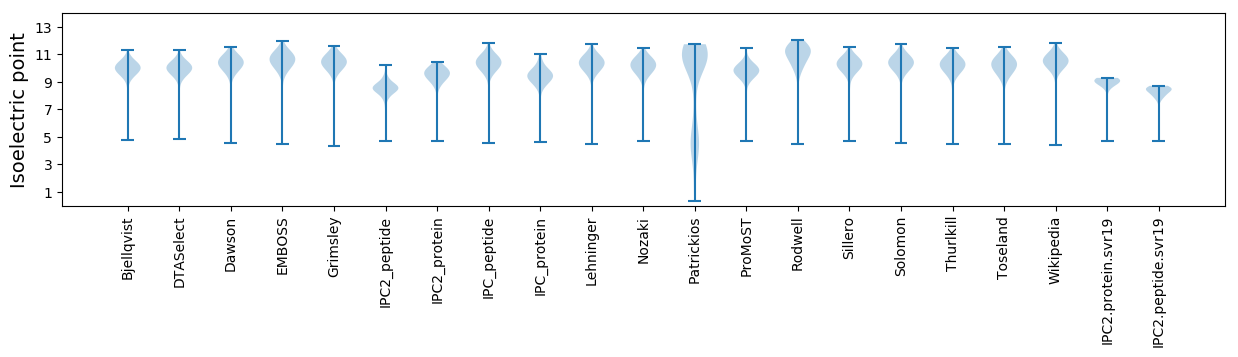
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|S5SQC5|S5SQC5_9PROT 50S ribosomal protein L15 OS=Candidatus Nasuia deltocephalinicola str. NAS-ALF OX=1343077 GN=rplO PE=3 SV=1
MM1 pKa = 6.74QKK3 pKa = 9.54EE4 pKa = 4.12RR5 pKa = 11.84KK6 pKa = 9.5RR7 pKa = 11.84FFLFNKK13 pKa = 8.85YY14 pKa = 8.52YY15 pKa = 10.85CKK17 pKa = 10.73LKK19 pKa = 10.68LLKK22 pKa = 10.5NQLNKK27 pKa = 10.28IIINKK32 pKa = 9.88SNTKK36 pKa = 9.76EE37 pKa = 4.05YY38 pKa = 10.76YY39 pKa = 10.05SEE41 pKa = 3.95LLKK44 pKa = 11.37VNFKK48 pKa = 10.01IQKK51 pKa = 9.84LPRR54 pKa = 11.84NSNGTRR60 pKa = 11.84IRR62 pKa = 11.84NRR64 pKa = 11.84CYY66 pKa = 8.66ITGRR70 pKa = 11.84PRR72 pKa = 11.84GVLRR76 pKa = 11.84LFGLSRR82 pKa = 11.84SIIRR86 pKa = 11.84DD87 pKa = 3.18IAFKK91 pKa = 10.76GYY93 pKa = 10.1IPGILKK99 pKa = 10.64SSWW102 pKa = 2.91
MM1 pKa = 6.74QKK3 pKa = 9.54EE4 pKa = 4.12RR5 pKa = 11.84KK6 pKa = 9.5RR7 pKa = 11.84FFLFNKK13 pKa = 8.85YY14 pKa = 8.52YY15 pKa = 10.85CKK17 pKa = 10.73LKK19 pKa = 10.68LLKK22 pKa = 10.5NQLNKK27 pKa = 10.28IIINKK32 pKa = 9.88SNTKK36 pKa = 9.76EE37 pKa = 4.05YY38 pKa = 10.76YY39 pKa = 10.05SEE41 pKa = 3.95LLKK44 pKa = 11.37VNFKK48 pKa = 10.01IQKK51 pKa = 9.84LPRR54 pKa = 11.84NSNGTRR60 pKa = 11.84IRR62 pKa = 11.84NRR64 pKa = 11.84CYY66 pKa = 8.66ITGRR70 pKa = 11.84PRR72 pKa = 11.84GVLRR76 pKa = 11.84LFGLSRR82 pKa = 11.84SIIRR86 pKa = 11.84DD87 pKa = 3.18IAFKK91 pKa = 10.76GYY93 pKa = 10.1IPGILKK99 pKa = 10.64SSWW102 pKa = 2.91
Molecular weight: 12.25 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
34211 |
33 |
1343 |
249.7 |
29.81 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
1.365 ± 0.157 | 1.377 ± 0.079 |
3.107 ± 0.182 | 3.391 ± 0.24 |
10.052 ± 0.552 | 3.622 ± 0.193 |
0.982 ± 0.064 | 14.685 ± 0.269 |
14.715 ± 0.466 | 10.459 ± 0.205 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.31 ± 0.081 | 11.923 ± 0.258 |
1.564 ± 0.106 | 1.225 ± 0.082 |
2.318 ± 0.115 | 6.302 ± 0.226 |
2.344 ± 0.137 | 2.616 ± 0.142 |
0.517 ± 0.053 | 6.127 ± 0.192 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
