
uncultured SUP05 cluster bacterium
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Gammaproteobacteria incertae sedis; Candidatus Thioglobus; environmental samples
Average proteome isoelectric point is 6.4
Get precalculated fractions of proteins
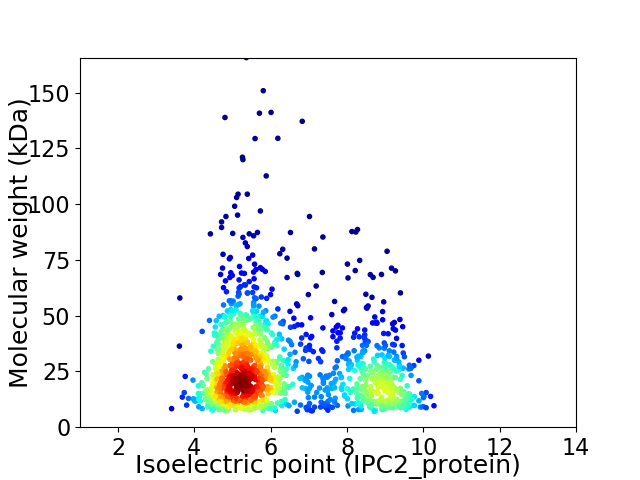
Virtual 2D-PAGE plot for 1283 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|D1KBD7|D1KBD7_9GAMM Protein-tyrosine-phosphatase OS=uncultured SUP05 cluster bacterium OX=655186 GN=Sup05_0233 PE=4 SV=1
MM1 pKa = 6.86VTTIDD6 pKa = 4.28ADD8 pKa = 4.0LQAMPSTTWSAWVKK22 pKa = 10.73PNGTSGWQVIFGMEE36 pKa = 3.84DD37 pKa = 3.34GGWDD41 pKa = 3.0RR42 pKa = 11.84FLILEE47 pKa = 4.4SGSLNVSMGHH57 pKa = 6.01TSNRR61 pKa = 11.84WQTGATLTNDD71 pKa = 3.07VWQHH75 pKa = 4.35VASIYY80 pKa = 11.12DD81 pKa = 3.4NGTMKK86 pKa = 10.56FYY88 pKa = 11.25LNGTEE93 pKa = 4.24YY94 pKa = 8.16TTGTDD99 pKa = 3.48EE100 pKa = 4.97GNHH103 pKa = 5.93SSTGKK108 pKa = 8.63FTIGGNQNGASNLYY122 pKa = 9.06VGRR125 pKa = 11.84VDD127 pKa = 5.83EE128 pKa = 4.26VAVWNEE134 pKa = 3.45ALTKK138 pKa = 10.76DD139 pKa = 4.51EE140 pKa = 5.31IVALYY145 pKa = 10.86NSGSPLLASSDD156 pKa = 3.68SGDD159 pKa = 3.65YY160 pKa = 10.78ASSGNLVGYY169 pKa = 7.94YY170 pKa = 10.06TMNSNDD176 pKa = 3.28GSGTSVTDD184 pKa = 3.58DD185 pKa = 3.58SGNSEE190 pKa = 4.18TATITGGSIWSTDD203 pKa = 3.02VVAITDD209 pKa = 3.69TTAPTLSSSTPADD222 pKa = 3.24NAASVAANANIVLNFSEE239 pKa = 4.5AVDD242 pKa = 3.87VEE244 pKa = 4.58SGKK247 pKa = 8.53ITIKK251 pKa = 9.69KK252 pKa = 8.47TSDD255 pKa = 2.89NSTVATIAVGDD266 pKa = 3.97AQVTGTGTTAITINPTNDD284 pKa = 2.9LSGGTEE290 pKa = 4.31YY291 pKa = 11.25YY292 pKa = 11.16VLIDD296 pKa = 3.42ATAFDD301 pKa = 4.95DD302 pKa = 4.58SSSNSYY308 pKa = 11.08AGISSTTALSFTTADD323 pKa = 3.52TTSPTISSTTVAADD337 pKa = 3.3NPTIDD342 pKa = 3.29VTFNEE347 pKa = 4.49AVFNATGGSGALQASDD363 pKa = 3.42FTISISGGTKK373 pKa = 8.81TLSSATPSSISISGNVYY390 pKa = 8.9TLGLDD395 pKa = 4.28LSGTANGSEE404 pKa = 4.61TITVVPSSSTAIYY417 pKa = 10.4DD418 pKa = 3.73GSDD421 pKa = 3.11NAASTSQSNNTVSLNSEE438 pKa = 4.33DD439 pKa = 3.9SSAPTLSSSSPTDD452 pKa = 3.32GAGSVAVTSNIVLTFSEE469 pKa = 4.8TVDD472 pKa = 3.56VEE474 pKa = 4.3SGDD477 pKa = 3.49IVLYY481 pKa = 10.57KK482 pKa = 10.58ADD484 pKa = 3.66GTVVEE489 pKa = 4.8TFSLPSAKK497 pKa = 9.0VTGTGSTTITINPTANLSSSTDD519 pKa = 3.43YY520 pKa = 11.04YY521 pKa = 11.22LQIAATAFDD530 pKa = 4.48DD531 pKa = 4.41SASNSYY537 pKa = 11.13AGITDD542 pKa = 3.63TTTLNFTTVGSDD554 pKa = 2.92PTTNKK559 pKa = 9.5TSPLQSKK566 pKa = 7.38QQ567 pKa = 3.12
MM1 pKa = 6.86VTTIDD6 pKa = 4.28ADD8 pKa = 4.0LQAMPSTTWSAWVKK22 pKa = 10.73PNGTSGWQVIFGMEE36 pKa = 3.84DD37 pKa = 3.34GGWDD41 pKa = 3.0RR42 pKa = 11.84FLILEE47 pKa = 4.4SGSLNVSMGHH57 pKa = 6.01TSNRR61 pKa = 11.84WQTGATLTNDD71 pKa = 3.07VWQHH75 pKa = 4.35VASIYY80 pKa = 11.12DD81 pKa = 3.4NGTMKK86 pKa = 10.56FYY88 pKa = 11.25LNGTEE93 pKa = 4.24YY94 pKa = 8.16TTGTDD99 pKa = 3.48EE100 pKa = 4.97GNHH103 pKa = 5.93SSTGKK108 pKa = 8.63FTIGGNQNGASNLYY122 pKa = 9.06VGRR125 pKa = 11.84VDD127 pKa = 5.83EE128 pKa = 4.26VAVWNEE134 pKa = 3.45ALTKK138 pKa = 10.76DD139 pKa = 4.51EE140 pKa = 5.31IVALYY145 pKa = 10.86NSGSPLLASSDD156 pKa = 3.68SGDD159 pKa = 3.65YY160 pKa = 10.78ASSGNLVGYY169 pKa = 7.94YY170 pKa = 10.06TMNSNDD176 pKa = 3.28GSGTSVTDD184 pKa = 3.58DD185 pKa = 3.58SGNSEE190 pKa = 4.18TATITGGSIWSTDD203 pKa = 3.02VVAITDD209 pKa = 3.69TTAPTLSSSTPADD222 pKa = 3.24NAASVAANANIVLNFSEE239 pKa = 4.5AVDD242 pKa = 3.87VEE244 pKa = 4.58SGKK247 pKa = 8.53ITIKK251 pKa = 9.69KK252 pKa = 8.47TSDD255 pKa = 2.89NSTVATIAVGDD266 pKa = 3.97AQVTGTGTTAITINPTNDD284 pKa = 2.9LSGGTEE290 pKa = 4.31YY291 pKa = 11.25YY292 pKa = 11.16VLIDD296 pKa = 3.42ATAFDD301 pKa = 4.95DD302 pKa = 4.58SSSNSYY308 pKa = 11.08AGISSTTALSFTTADD323 pKa = 3.52TTSPTISSTTVAADD337 pKa = 3.3NPTIDD342 pKa = 3.29VTFNEE347 pKa = 4.49AVFNATGGSGALQASDD363 pKa = 3.42FTISISGGTKK373 pKa = 8.81TLSSATPSSISISGNVYY390 pKa = 8.9TLGLDD395 pKa = 4.28LSGTANGSEE404 pKa = 4.61TITVVPSSSTAIYY417 pKa = 10.4DD418 pKa = 3.73GSDD421 pKa = 3.11NAASTSQSNNTVSLNSEE438 pKa = 4.33DD439 pKa = 3.9SSAPTLSSSSPTDD452 pKa = 3.32GAGSVAVTSNIVLTFSEE469 pKa = 4.8TVDD472 pKa = 3.56VEE474 pKa = 4.3SGDD477 pKa = 3.49IVLYY481 pKa = 10.57KK482 pKa = 10.58ADD484 pKa = 3.66GTVVEE489 pKa = 4.8TFSLPSAKK497 pKa = 9.0VTGTGSTTITINPTANLSSSTDD519 pKa = 3.43YY520 pKa = 11.04YY521 pKa = 11.22LQIAATAFDD530 pKa = 4.48DD531 pKa = 4.41SASNSYY537 pKa = 11.13AGITDD542 pKa = 3.63TTTLNFTTVGSDD554 pKa = 2.92PTTNKK559 pKa = 9.5TSPLQSKK566 pKa = 7.38QQ567 pKa = 3.12
Molecular weight: 57.93 kDa
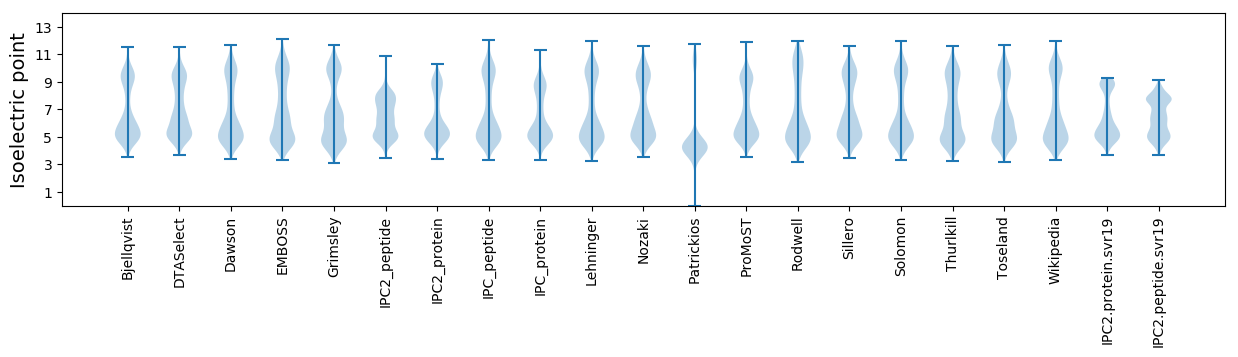
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|D1KDM2|D1KDM2_9GAMM DUF4396 domain-containing protein OS=uncultured SUP05 cluster bacterium OX=655186 GN=Sup05_0638 PE=4 SV=1
MM1 pKa = 6.99AQVVKK6 pKa = 10.61RR7 pKa = 11.84KK8 pKa = 6.54PTSPGRR14 pKa = 11.84RR15 pKa = 11.84FVVSIVDD22 pKa = 3.49KK23 pKa = 10.48DD24 pKa = 3.63LHH26 pKa = 7.05KK27 pKa = 10.53GAPYY31 pKa = 10.82APLTEE36 pKa = 4.29SKK38 pKa = 10.93NRR40 pKa = 11.84ISGRR44 pKa = 11.84NNRR47 pKa = 11.84GKK49 pKa = 8.86ITVRR53 pKa = 11.84HH54 pKa = 6.09KK55 pKa = 11.19GGGHH59 pKa = 5.21KK60 pKa = 9.8RR61 pKa = 11.84RR62 pKa = 11.84YY63 pKa = 7.88RR64 pKa = 11.84TIDD67 pKa = 3.41FKK69 pKa = 11.0RR70 pKa = 11.84IKK72 pKa = 10.41DD73 pKa = 4.34DD74 pKa = 3.2IPATVEE80 pKa = 3.74RR81 pKa = 11.84LEE83 pKa = 4.03YY84 pKa = 10.85DD85 pKa = 3.48PNRR88 pKa = 11.84SANIALVLYY97 pKa = 10.67ADD99 pKa = 4.34GEE101 pKa = 4.34RR102 pKa = 11.84TYY104 pKa = 10.63IIAPNGLKK112 pKa = 10.73VGDD115 pKa = 4.03TVVSGTSVAIQAGNVMPCANIPLGSVIHH143 pKa = 6.55CIEE146 pKa = 4.45LKK148 pKa = 9.83PGKK151 pKa = 8.83GAQMARR157 pKa = 11.84SAGTSAQLIAKK168 pKa = 8.31EE169 pKa = 4.17GTYY172 pKa = 9.08VTLRR176 pKa = 11.84LRR178 pKa = 11.84SGEE181 pKa = 3.97VRR183 pKa = 11.84KK184 pKa = 10.26VLAEE188 pKa = 3.87CRR190 pKa = 11.84ATIGEE195 pKa = 4.22VSKK198 pKa = 11.08KK199 pKa = 7.95EE200 pKa = 3.6HH201 pKa = 6.15SLRR204 pKa = 11.84KK205 pKa = 9.46LGKK208 pKa = 10.11AGATRR213 pKa = 11.84WRR215 pKa = 11.84GVRR218 pKa = 11.84PTVRR222 pKa = 11.84GVVMNPVDD230 pKa = 3.77HH231 pKa = 6.67PHH233 pKa = 6.77GGGEE237 pKa = 4.38GKK239 pKa = 9.02TSGGRR244 pKa = 11.84HH245 pKa = 5.43PVSPWGTPTKK255 pKa = 10.39GYY257 pKa = 7.85KK258 pKa = 8.71TRR260 pKa = 11.84SNKK263 pKa = 8.79RR264 pKa = 11.84TDD266 pKa = 3.07KK267 pKa = 11.11LIVRR271 pKa = 11.84RR272 pKa = 11.84RR273 pKa = 11.84NKK275 pKa = 10.26
MM1 pKa = 6.99AQVVKK6 pKa = 10.61RR7 pKa = 11.84KK8 pKa = 6.54PTSPGRR14 pKa = 11.84RR15 pKa = 11.84FVVSIVDD22 pKa = 3.49KK23 pKa = 10.48DD24 pKa = 3.63LHH26 pKa = 7.05KK27 pKa = 10.53GAPYY31 pKa = 10.82APLTEE36 pKa = 4.29SKK38 pKa = 10.93NRR40 pKa = 11.84ISGRR44 pKa = 11.84NNRR47 pKa = 11.84GKK49 pKa = 8.86ITVRR53 pKa = 11.84HH54 pKa = 6.09KK55 pKa = 11.19GGGHH59 pKa = 5.21KK60 pKa = 9.8RR61 pKa = 11.84RR62 pKa = 11.84YY63 pKa = 7.88RR64 pKa = 11.84TIDD67 pKa = 3.41FKK69 pKa = 11.0RR70 pKa = 11.84IKK72 pKa = 10.41DD73 pKa = 4.34DD74 pKa = 3.2IPATVEE80 pKa = 3.74RR81 pKa = 11.84LEE83 pKa = 4.03YY84 pKa = 10.85DD85 pKa = 3.48PNRR88 pKa = 11.84SANIALVLYY97 pKa = 10.67ADD99 pKa = 4.34GEE101 pKa = 4.34RR102 pKa = 11.84TYY104 pKa = 10.63IIAPNGLKK112 pKa = 10.73VGDD115 pKa = 4.03TVVSGTSVAIQAGNVMPCANIPLGSVIHH143 pKa = 6.55CIEE146 pKa = 4.45LKK148 pKa = 9.83PGKK151 pKa = 8.83GAQMARR157 pKa = 11.84SAGTSAQLIAKK168 pKa = 8.31EE169 pKa = 4.17GTYY172 pKa = 9.08VTLRR176 pKa = 11.84LRR178 pKa = 11.84SGEE181 pKa = 3.97VRR183 pKa = 11.84KK184 pKa = 10.26VLAEE188 pKa = 3.87CRR190 pKa = 11.84ATIGEE195 pKa = 4.22VSKK198 pKa = 11.08KK199 pKa = 7.95EE200 pKa = 3.6HH201 pKa = 6.15SLRR204 pKa = 11.84KK205 pKa = 9.46LGKK208 pKa = 10.11AGATRR213 pKa = 11.84WRR215 pKa = 11.84GVRR218 pKa = 11.84PTVRR222 pKa = 11.84GVVMNPVDD230 pKa = 3.77HH231 pKa = 6.67PHH233 pKa = 6.77GGGEE237 pKa = 4.38GKK239 pKa = 9.02TSGGRR244 pKa = 11.84HH245 pKa = 5.43PVSPWGTPTKK255 pKa = 10.39GYY257 pKa = 7.85KK258 pKa = 8.71TRR260 pKa = 11.84SNKK263 pKa = 8.79RR264 pKa = 11.84TDD266 pKa = 3.07KK267 pKa = 11.11LIVRR271 pKa = 11.84RR272 pKa = 11.84RR273 pKa = 11.84NKK275 pKa = 10.26
Molecular weight: 30.03 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
338736 |
67 |
1499 |
264.0 |
29.41 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.463 ± 0.069 | 1.007 ± 0.028 |
6.085 ± 0.055 | 5.911 ± 0.055 |
4.267 ± 0.049 | 6.806 ± 0.064 |
2.232 ± 0.038 | 7.615 ± 0.057 |
7.143 ± 0.075 | 9.911 ± 0.085 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.712 ± 0.04 | 4.94 ± 0.053 |
3.388 ± 0.04 | 4.013 ± 0.05 |
3.757 ± 0.047 | 6.724 ± 0.059 |
5.313 ± 0.047 | 6.719 ± 0.057 |
0.986 ± 0.028 | 3.008 ± 0.034 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
