
Larkinella punicea
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Cytophagia; Cytophagales; Cytophagaceae; Larkinella
Average proteome isoelectric point is 6.9
Get precalculated fractions of proteins
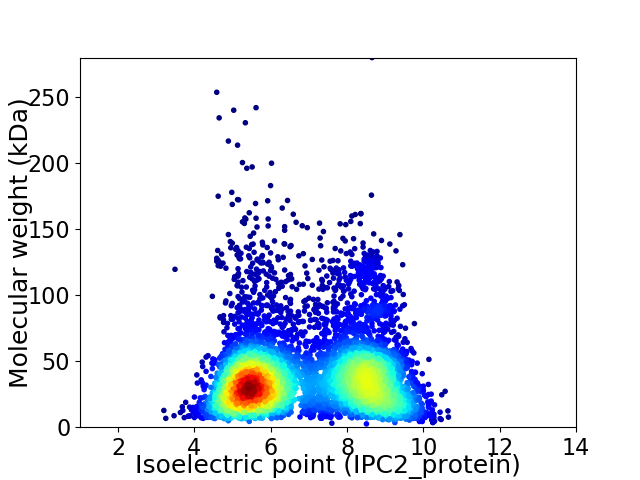
Virtual 2D-PAGE plot for 6294 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A368JVB2|A0A368JVB2_9BACT AraC family transcriptional regulator OS=Larkinella punicea OX=2315727 GN=DUE52_06505 PE=4 SV=1
MM1 pKa = 7.43SLLAVMGMLVSIPPAYY17 pKa = 10.05AQSCYY22 pKa = 11.15APISGTAVQVGGVISATVCAGPLCLGTGTYY52 pKa = 10.75NEE54 pKa = 4.28LQKK57 pKa = 9.83ITDD60 pKa = 3.47NSLEE64 pKa = 4.04TFANINTTIAAQIAQGISVKK84 pKa = 9.38RR85 pKa = 11.84TNGTYY90 pKa = 9.57TGEE93 pKa = 4.11NVVGFILSRR102 pKa = 11.84SALVDD107 pKa = 3.18VSVQSSVTIRR117 pKa = 11.84TYY119 pKa = 11.5LNGNPVSAAQPLNALVSGSALGSVEE144 pKa = 4.33KK145 pKa = 10.13MYY147 pKa = 10.11ATFTTNQEE155 pKa = 3.73FDD157 pKa = 3.54EE158 pKa = 4.42VRR160 pKa = 11.84LNTSLLSAEE169 pKa = 4.28VFSNLRR175 pKa = 11.84IFAAVAFPTSCSTPVSTTACNDD197 pKa = 3.7YY198 pKa = 10.9ISGLGTDD205 pKa = 3.15ATYY208 pKa = 10.87YY209 pKa = 11.22GSVTCAACSLTDD221 pKa = 3.65RR222 pKa = 11.84ANLIDD227 pKa = 4.52GNNANYY233 pKa = 10.61AVLTPTAGVASEE245 pKa = 4.34ISVSVIDD252 pKa = 3.67TKK254 pKa = 11.13KK255 pKa = 10.29KK256 pKa = 10.12YY257 pKa = 9.94KK258 pKa = 10.7AGDD261 pKa = 3.22RR262 pKa = 11.84AGFVIAKK269 pKa = 8.84NQVNALLALNALSTITIEE287 pKa = 4.22TYY289 pKa = 11.0LFGTLQEE296 pKa = 4.56AKK298 pKa = 10.73SPGDD302 pKa = 3.35PTNVVGLSLLAGSNTIPKK320 pKa = 9.59QKK322 pKa = 10.65LGFTTTLDD330 pKa = 3.62FNEE333 pKa = 3.4VRR335 pKa = 11.84IRR337 pKa = 11.84INSLATANVGAIRR350 pKa = 11.84LYY352 pKa = 10.77GAFVEE357 pKa = 5.2ATTCTNCEE365 pKa = 4.02TLLNNAGTGKK375 pKa = 10.56YY376 pKa = 9.92NGAIVTGFQSLGLPWTGVHH395 pKa = 6.68GLGLHH400 pKa = 6.89ALTNSGNAVTSTPTDD415 pKa = 3.28DD416 pKa = 3.45FATYY420 pKa = 9.18TSVVGVLGAGLQFTVEE436 pKa = 3.85NDD438 pKa = 3.54GTDD441 pKa = 3.86FTANTTFAGFHH452 pKa = 5.86ISKK455 pKa = 8.79TGSLIALGLLDD466 pKa = 5.87AITIKK471 pKa = 10.69VYY473 pKa = 11.1NGTTLVDD480 pKa = 3.88EE481 pKa = 4.57STSASLLGASLISGTTTRR499 pKa = 11.84SVVGFYY505 pKa = 10.49PDD507 pKa = 3.14AAFDD511 pKa = 5.01RR512 pKa = 11.84IQLIVNEE519 pKa = 4.63GLLSADD525 pKa = 3.52LAGDD529 pKa = 3.55YY530 pKa = 10.72LIYY533 pKa = 10.56DD534 pKa = 3.95AFVIEE539 pKa = 4.97DD540 pKa = 3.76ADD542 pKa = 4.03GDD544 pKa = 4.42GVPDD548 pKa = 4.06CQDD551 pKa = 2.81VCGTGTGNDD560 pKa = 3.95LLDD563 pKa = 4.11FDD565 pKa = 5.76GDD567 pKa = 4.77GIPDD571 pKa = 3.71TCDD574 pKa = 3.65PDD576 pKa = 5.03DD577 pKa = 5.85DD578 pKa = 5.14NDD580 pKa = 5.16GIPDD584 pKa = 3.71TVEE587 pKa = 3.94NPEE590 pKa = 4.14GDD592 pKa = 3.82PNRR595 pKa = 11.84DD596 pKa = 2.98TDD598 pKa = 4.06GDD600 pKa = 4.07SSPDD604 pKa = 3.92LRR606 pKa = 11.84DD607 pKa = 3.5LDD609 pKa = 4.27SDD611 pKa = 3.78NDD613 pKa = 4.74GIPDD617 pKa = 3.89SVEE620 pKa = 3.88KK621 pKa = 11.09GPDD624 pKa = 3.25GNNPSNTDD632 pKa = 3.06GTDD635 pKa = 3.15VPDD638 pKa = 3.92YY639 pKa = 10.81RR640 pKa = 11.84DD641 pKa = 3.8PDD643 pKa = 3.78SDD645 pKa = 3.91NDD647 pKa = 4.73GIPDD651 pKa = 3.68SVEE654 pKa = 3.76AGPNGDD660 pKa = 3.82EE661 pKa = 4.61PRR663 pKa = 11.84NTDD666 pKa = 3.11GDD668 pKa = 4.27ALPDD672 pKa = 3.9YY673 pKa = 11.2LDD675 pKa = 5.06LDD677 pKa = 4.06SDD679 pKa = 3.94NDD681 pKa = 4.73GILDD685 pKa = 3.95TVEE688 pKa = 4.63KK689 pKa = 10.8GSDD692 pKa = 3.41GNNPTNSDD700 pKa = 3.85TNTGSNPDD708 pKa = 3.62GRR710 pKa = 11.84PDD712 pKa = 4.83YY713 pKa = 11.01IDD715 pKa = 5.67LDD717 pKa = 4.07ADD719 pKa = 3.53NDD721 pKa = 4.74GINDD725 pKa = 3.8VIEE728 pKa = 4.53ANPNGSPTKK737 pKa = 10.36DD738 pKa = 3.4SNGDD742 pKa = 3.72GIADD746 pKa = 4.11GTPGATGIPASAGSGLTPPDD766 pKa = 3.38TDD768 pKa = 4.54GDD770 pKa = 4.2TKK772 pKa = 11.21PDD774 pKa = 3.64FQDD777 pKa = 3.15IDD779 pKa = 3.73TDD781 pKa = 3.95GDD783 pKa = 4.4GIPDD787 pKa = 4.33LYY789 pKa = 11.09EE790 pKa = 4.74SGIPNPNALDD800 pKa = 3.75TNKK803 pKa = 10.9DD804 pKa = 3.66GIIDD808 pKa = 4.11DD809 pKa = 4.93ASEE812 pKa = 4.1TDD814 pKa = 3.3NDD816 pKa = 5.44GIPTVNAPAAVVDD829 pKa = 4.59GANGTYY835 pKa = 10.88GDD837 pKa = 4.53ANSPALPNNDD847 pKa = 2.17GDD849 pKa = 4.31AYY851 pKa = 10.21PNYY854 pKa = 10.04RR855 pKa = 11.84DD856 pKa = 4.59LDD858 pKa = 3.84SDD860 pKa = 3.75NDD862 pKa = 4.86GITDD866 pKa = 3.88TVEE869 pKa = 3.92KK870 pKa = 10.57GIDD873 pKa = 3.53GANPTNTDD881 pKa = 2.79GDD883 pKa = 4.53LLPDD887 pKa = 3.94YY888 pKa = 11.18LDD890 pKa = 4.96LDD892 pKa = 4.15SDD894 pKa = 3.93NDD896 pKa = 4.59GINDD900 pKa = 3.5VVEE903 pKa = 4.39ATGSPTNDD911 pKa = 3.15SNGDD915 pKa = 3.84GIADD919 pKa = 4.11GTPGATGIPSSAGSGVTPADD939 pKa = 3.84ADD941 pKa = 3.76NDD943 pKa = 3.75GSPNYY948 pKa = 9.88RR949 pKa = 11.84DD950 pKa = 4.12LDD952 pKa = 3.7SDD954 pKa = 3.83NDD956 pKa = 4.14SKK958 pKa = 11.71KK959 pKa = 10.71DD960 pKa = 3.56LYY962 pKa = 11.19EE963 pKa = 4.8SGIPNPASLDD973 pKa = 3.76TNGDD977 pKa = 3.84GVVDD981 pKa = 4.17AADD984 pKa = 3.77GTDD987 pKa = 3.45NDD989 pKa = 5.84GIPQTVDD996 pKa = 3.29GEE998 pKa = 4.37PNAYY1002 pKa = 9.73GDD1004 pKa = 4.13SGNPTLPNEE1013 pKa = 4.6DD1014 pKa = 3.23TDD1016 pKa = 4.08EE1017 pKa = 4.65NPDD1020 pKa = 3.91YY1021 pKa = 10.73IDD1023 pKa = 3.81PAGVDD1028 pKa = 4.19LYY1030 pKa = 9.47PTLSMPDD1037 pKa = 3.39RR1038 pKa = 11.84LYY1040 pKa = 11.13GASGGAKK1047 pKa = 8.42NLKK1050 pKa = 8.04VQVYY1054 pKa = 10.17NLIPATNTNGTITLTISRR1072 pKa = 11.84PSPFYY1077 pKa = 11.07TMALSTAEE1085 pKa = 4.07ALQDD1089 pKa = 3.23QWILTTEE1096 pKa = 3.94TLLFKK1101 pKa = 10.63LVSKK1105 pKa = 9.73TGVTIAGSSFNEE1117 pKa = 3.64IEE1119 pKa = 4.27MVLTASGSAAPGLANLNINILAGSGGEE1146 pKa = 4.1TNTANNVASAQITIEE1161 pKa = 3.9
MM1 pKa = 7.43SLLAVMGMLVSIPPAYY17 pKa = 10.05AQSCYY22 pKa = 11.15APISGTAVQVGGVISATVCAGPLCLGTGTYY52 pKa = 10.75NEE54 pKa = 4.28LQKK57 pKa = 9.83ITDD60 pKa = 3.47NSLEE64 pKa = 4.04TFANINTTIAAQIAQGISVKK84 pKa = 9.38RR85 pKa = 11.84TNGTYY90 pKa = 9.57TGEE93 pKa = 4.11NVVGFILSRR102 pKa = 11.84SALVDD107 pKa = 3.18VSVQSSVTIRR117 pKa = 11.84TYY119 pKa = 11.5LNGNPVSAAQPLNALVSGSALGSVEE144 pKa = 4.33KK145 pKa = 10.13MYY147 pKa = 10.11ATFTTNQEE155 pKa = 3.73FDD157 pKa = 3.54EE158 pKa = 4.42VRR160 pKa = 11.84LNTSLLSAEE169 pKa = 4.28VFSNLRR175 pKa = 11.84IFAAVAFPTSCSTPVSTTACNDD197 pKa = 3.7YY198 pKa = 10.9ISGLGTDD205 pKa = 3.15ATYY208 pKa = 10.87YY209 pKa = 11.22GSVTCAACSLTDD221 pKa = 3.65RR222 pKa = 11.84ANLIDD227 pKa = 4.52GNNANYY233 pKa = 10.61AVLTPTAGVASEE245 pKa = 4.34ISVSVIDD252 pKa = 3.67TKK254 pKa = 11.13KK255 pKa = 10.29KK256 pKa = 10.12YY257 pKa = 9.94KK258 pKa = 10.7AGDD261 pKa = 3.22RR262 pKa = 11.84AGFVIAKK269 pKa = 8.84NQVNALLALNALSTITIEE287 pKa = 4.22TYY289 pKa = 11.0LFGTLQEE296 pKa = 4.56AKK298 pKa = 10.73SPGDD302 pKa = 3.35PTNVVGLSLLAGSNTIPKK320 pKa = 9.59QKK322 pKa = 10.65LGFTTTLDD330 pKa = 3.62FNEE333 pKa = 3.4VRR335 pKa = 11.84IRR337 pKa = 11.84INSLATANVGAIRR350 pKa = 11.84LYY352 pKa = 10.77GAFVEE357 pKa = 5.2ATTCTNCEE365 pKa = 4.02TLLNNAGTGKK375 pKa = 10.56YY376 pKa = 9.92NGAIVTGFQSLGLPWTGVHH395 pKa = 6.68GLGLHH400 pKa = 6.89ALTNSGNAVTSTPTDD415 pKa = 3.28DD416 pKa = 3.45FATYY420 pKa = 9.18TSVVGVLGAGLQFTVEE436 pKa = 3.85NDD438 pKa = 3.54GTDD441 pKa = 3.86FTANTTFAGFHH452 pKa = 5.86ISKK455 pKa = 8.79TGSLIALGLLDD466 pKa = 5.87AITIKK471 pKa = 10.69VYY473 pKa = 11.1NGTTLVDD480 pKa = 3.88EE481 pKa = 4.57STSASLLGASLISGTTTRR499 pKa = 11.84SVVGFYY505 pKa = 10.49PDD507 pKa = 3.14AAFDD511 pKa = 5.01RR512 pKa = 11.84IQLIVNEE519 pKa = 4.63GLLSADD525 pKa = 3.52LAGDD529 pKa = 3.55YY530 pKa = 10.72LIYY533 pKa = 10.56DD534 pKa = 3.95AFVIEE539 pKa = 4.97DD540 pKa = 3.76ADD542 pKa = 4.03GDD544 pKa = 4.42GVPDD548 pKa = 4.06CQDD551 pKa = 2.81VCGTGTGNDD560 pKa = 3.95LLDD563 pKa = 4.11FDD565 pKa = 5.76GDD567 pKa = 4.77GIPDD571 pKa = 3.71TCDD574 pKa = 3.65PDD576 pKa = 5.03DD577 pKa = 5.85DD578 pKa = 5.14NDD580 pKa = 5.16GIPDD584 pKa = 3.71TVEE587 pKa = 3.94NPEE590 pKa = 4.14GDD592 pKa = 3.82PNRR595 pKa = 11.84DD596 pKa = 2.98TDD598 pKa = 4.06GDD600 pKa = 4.07SSPDD604 pKa = 3.92LRR606 pKa = 11.84DD607 pKa = 3.5LDD609 pKa = 4.27SDD611 pKa = 3.78NDD613 pKa = 4.74GIPDD617 pKa = 3.89SVEE620 pKa = 3.88KK621 pKa = 11.09GPDD624 pKa = 3.25GNNPSNTDD632 pKa = 3.06GTDD635 pKa = 3.15VPDD638 pKa = 3.92YY639 pKa = 10.81RR640 pKa = 11.84DD641 pKa = 3.8PDD643 pKa = 3.78SDD645 pKa = 3.91NDD647 pKa = 4.73GIPDD651 pKa = 3.68SVEE654 pKa = 3.76AGPNGDD660 pKa = 3.82EE661 pKa = 4.61PRR663 pKa = 11.84NTDD666 pKa = 3.11GDD668 pKa = 4.27ALPDD672 pKa = 3.9YY673 pKa = 11.2LDD675 pKa = 5.06LDD677 pKa = 4.06SDD679 pKa = 3.94NDD681 pKa = 4.73GILDD685 pKa = 3.95TVEE688 pKa = 4.63KK689 pKa = 10.8GSDD692 pKa = 3.41GNNPTNSDD700 pKa = 3.85TNTGSNPDD708 pKa = 3.62GRR710 pKa = 11.84PDD712 pKa = 4.83YY713 pKa = 11.01IDD715 pKa = 5.67LDD717 pKa = 4.07ADD719 pKa = 3.53NDD721 pKa = 4.74GINDD725 pKa = 3.8VIEE728 pKa = 4.53ANPNGSPTKK737 pKa = 10.36DD738 pKa = 3.4SNGDD742 pKa = 3.72GIADD746 pKa = 4.11GTPGATGIPASAGSGLTPPDD766 pKa = 3.38TDD768 pKa = 4.54GDD770 pKa = 4.2TKK772 pKa = 11.21PDD774 pKa = 3.64FQDD777 pKa = 3.15IDD779 pKa = 3.73TDD781 pKa = 3.95GDD783 pKa = 4.4GIPDD787 pKa = 4.33LYY789 pKa = 11.09EE790 pKa = 4.74SGIPNPNALDD800 pKa = 3.75TNKK803 pKa = 10.9DD804 pKa = 3.66GIIDD808 pKa = 4.11DD809 pKa = 4.93ASEE812 pKa = 4.1TDD814 pKa = 3.3NDD816 pKa = 5.44GIPTVNAPAAVVDD829 pKa = 4.59GANGTYY835 pKa = 10.88GDD837 pKa = 4.53ANSPALPNNDD847 pKa = 2.17GDD849 pKa = 4.31AYY851 pKa = 10.21PNYY854 pKa = 10.04RR855 pKa = 11.84DD856 pKa = 4.59LDD858 pKa = 3.84SDD860 pKa = 3.75NDD862 pKa = 4.86GITDD866 pKa = 3.88TVEE869 pKa = 3.92KK870 pKa = 10.57GIDD873 pKa = 3.53GANPTNTDD881 pKa = 2.79GDD883 pKa = 4.53LLPDD887 pKa = 3.94YY888 pKa = 11.18LDD890 pKa = 4.96LDD892 pKa = 4.15SDD894 pKa = 3.93NDD896 pKa = 4.59GINDD900 pKa = 3.5VVEE903 pKa = 4.39ATGSPTNDD911 pKa = 3.15SNGDD915 pKa = 3.84GIADD919 pKa = 4.11GTPGATGIPSSAGSGVTPADD939 pKa = 3.84ADD941 pKa = 3.76NDD943 pKa = 3.75GSPNYY948 pKa = 9.88RR949 pKa = 11.84DD950 pKa = 4.12LDD952 pKa = 3.7SDD954 pKa = 3.83NDD956 pKa = 4.14SKK958 pKa = 11.71KK959 pKa = 10.71DD960 pKa = 3.56LYY962 pKa = 11.19EE963 pKa = 4.8SGIPNPASLDD973 pKa = 3.76TNGDD977 pKa = 3.84GVVDD981 pKa = 4.17AADD984 pKa = 3.77GTDD987 pKa = 3.45NDD989 pKa = 5.84GIPQTVDD996 pKa = 3.29GEE998 pKa = 4.37PNAYY1002 pKa = 9.73GDD1004 pKa = 4.13SGNPTLPNEE1013 pKa = 4.6DD1014 pKa = 3.23TDD1016 pKa = 4.08EE1017 pKa = 4.65NPDD1020 pKa = 3.91YY1021 pKa = 10.73IDD1023 pKa = 3.81PAGVDD1028 pKa = 4.19LYY1030 pKa = 9.47PTLSMPDD1037 pKa = 3.39RR1038 pKa = 11.84LYY1040 pKa = 11.13GASGGAKK1047 pKa = 8.42NLKK1050 pKa = 8.04VQVYY1054 pKa = 10.17NLIPATNTNGTITLTISRR1072 pKa = 11.84PSPFYY1077 pKa = 11.07TMALSTAEE1085 pKa = 4.07ALQDD1089 pKa = 3.23QWILTTEE1096 pKa = 3.94TLLFKK1101 pKa = 10.63LVSKK1105 pKa = 9.73TGVTIAGSSFNEE1117 pKa = 3.64IEE1119 pKa = 4.27MVLTASGSAAPGLANLNINILAGSGGEE1146 pKa = 4.1TNTANNVASAQITIEE1161 pKa = 3.9
Molecular weight: 119.49 kDa
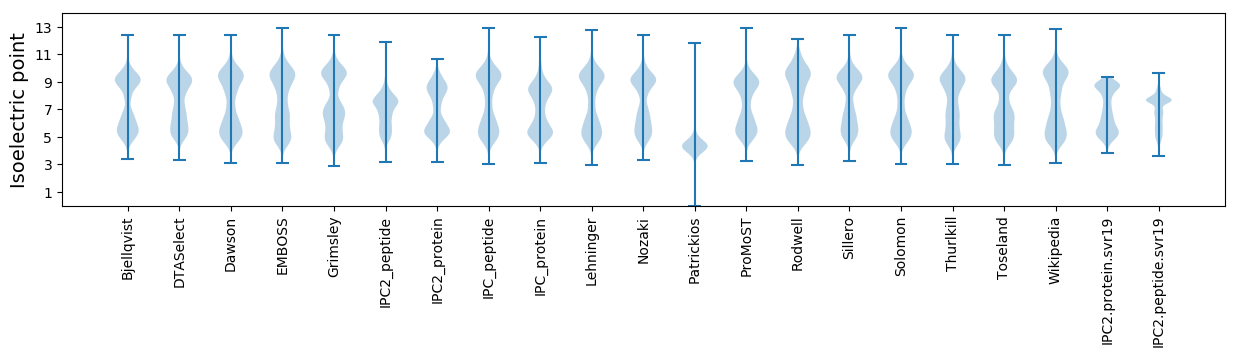
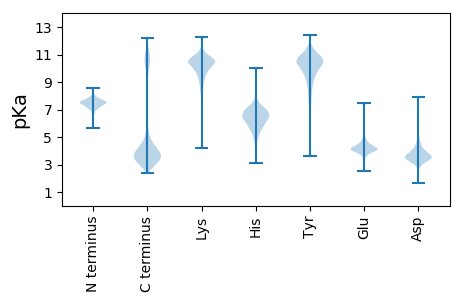
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A368JGN6|A0A368JGN6_9BACT 3-oxoacyl-[acyl-carrier-protein] synthase 3 OS=Larkinella punicea OX=2315727 GN=fabH PE=3 SV=1
MM1 pKa = 7.64PKK3 pKa = 10.07SLSFRR8 pKa = 11.84SGLGSVLFWSVISAAFIGPGTVTTCAMAGSRR39 pKa = 11.84FGLDD43 pKa = 3.69LLWALTFSTIGTIVLQEE60 pKa = 3.5AAARR64 pKa = 11.84ITIASGLSLGQVVARR79 pKa = 11.84SYY81 pKa = 10.1GTGSRR86 pKa = 11.84WIAGALLFSIVLGCAAYY103 pKa = 10.05QAGNILGAVAGLSMILPVSPVWLTAGIGLVCIFLLWQGSTRR144 pKa = 11.84LIANFLGVIVFVMGVAFAYY163 pKa = 10.18VATRR167 pKa = 11.84TDD169 pKa = 2.93VSAGMFVKK177 pKa = 10.74ALVVPVATADD187 pKa = 3.66SFVLVIGLIGTTIVPYY203 pKa = 10.94NLFLASGLGKK213 pKa = 10.22GQSLAEE219 pKa = 4.01MRR221 pKa = 11.84WGIGLAVLIGGLISMAILVTGTLVTGEE248 pKa = 4.75FSFQQVADD256 pKa = 3.84ALANRR261 pKa = 11.84VGGWSRR267 pKa = 11.84RR268 pKa = 11.84LFAFGLFAAGFTSALTAPMAAAVTSQSLLGWAGSSWRR305 pKa = 11.84YY306 pKa = 8.62RR307 pKa = 11.84LVWLGVMGVGLMFGLMNARR326 pKa = 11.84PVPVIIVVQAINGVLLPVVTIFLFRR351 pKa = 11.84AVNDD355 pKa = 3.48KK356 pKa = 11.05KK357 pKa = 10.96LIPEE361 pKa = 4.75AYY363 pKa = 9.81RR364 pKa = 11.84NSGLQNLAMLVVVAVSLLFGGWNVWLAVQNWLKK397 pKa = 11.12
MM1 pKa = 7.64PKK3 pKa = 10.07SLSFRR8 pKa = 11.84SGLGSVLFWSVISAAFIGPGTVTTCAMAGSRR39 pKa = 11.84FGLDD43 pKa = 3.69LLWALTFSTIGTIVLQEE60 pKa = 3.5AAARR64 pKa = 11.84ITIASGLSLGQVVARR79 pKa = 11.84SYY81 pKa = 10.1GTGSRR86 pKa = 11.84WIAGALLFSIVLGCAAYY103 pKa = 10.05QAGNILGAVAGLSMILPVSPVWLTAGIGLVCIFLLWQGSTRR144 pKa = 11.84LIANFLGVIVFVMGVAFAYY163 pKa = 10.18VATRR167 pKa = 11.84TDD169 pKa = 2.93VSAGMFVKK177 pKa = 10.74ALVVPVATADD187 pKa = 3.66SFVLVIGLIGTTIVPYY203 pKa = 10.94NLFLASGLGKK213 pKa = 10.22GQSLAEE219 pKa = 4.01MRR221 pKa = 11.84WGIGLAVLIGGLISMAILVTGTLVTGEE248 pKa = 4.75FSFQQVADD256 pKa = 3.84ALANRR261 pKa = 11.84VGGWSRR267 pKa = 11.84RR268 pKa = 11.84LFAFGLFAAGFTSALTAPMAAAVTSQSLLGWAGSSWRR305 pKa = 11.84YY306 pKa = 8.62RR307 pKa = 11.84LVWLGVMGVGLMFGLMNARR326 pKa = 11.84PVPVIIVVQAINGVLLPVVTIFLFRR351 pKa = 11.84AVNDD355 pKa = 3.48KK356 pKa = 11.05KK357 pKa = 10.96LIPEE361 pKa = 4.75AYY363 pKa = 9.81RR364 pKa = 11.84NSGLQNLAMLVVVAVSLLFGGWNVWLAVQNWLKK397 pKa = 11.12
Molecular weight: 41.61 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2371370 |
21 |
2512 |
376.8 |
42.09 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.941 ± 0.028 | 0.717 ± 0.009 |
5.236 ± 0.024 | 5.51 ± 0.027 |
4.672 ± 0.021 | 7.223 ± 0.029 |
1.815 ± 0.016 | 5.862 ± 0.026 |
5.449 ± 0.028 | 10.058 ± 0.035 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.071 ± 0.015 | 4.731 ± 0.024 |
4.639 ± 0.022 | 4.271 ± 0.02 |
5.3 ± 0.024 | 6.13 ± 0.024 |
6.313 ± 0.032 | 6.913 ± 0.022 |
1.363 ± 0.011 | 3.786 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
