
Ectocarpus sp. CCAP 1310/34
Taxonomy: cellular organisms; Eukaryota; Sar; Stramenopiles; Ochrophyta; PX clade; Phaeophyceae; Ectocarpales; Ectocarpaceae; Ectocarpus; unclassified Ectocarpus
Average proteome isoelectric point is 6.89
Get precalculated fractions of proteins
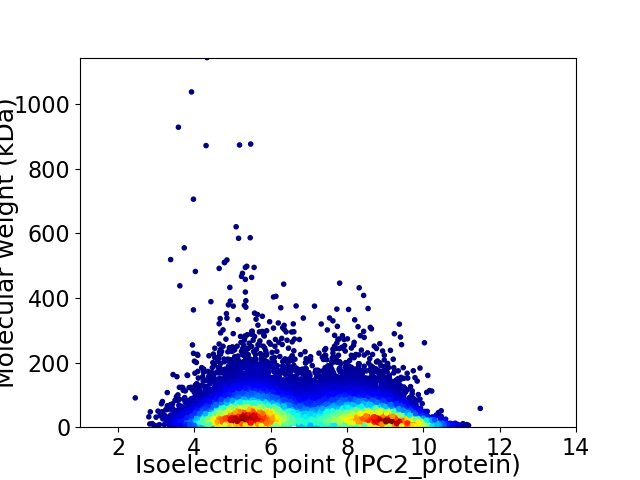
Virtual 2D-PAGE plot for 25330 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6H5JCI3|A0A6H5JCI3_9PHAE Uncharacterized protein (Fragment) OS=Ectocarpus sp. CCAP 1310/34 OX=867726 GN=ESUBFT_1007_7 PE=4 SV=1
MM1 pKa = 7.66SLEE4 pKa = 4.19FRR6 pKa = 11.84LDD8 pKa = 3.2LCGYY12 pKa = 10.14CDD14 pKa = 3.55TYY16 pKa = 11.96NNGSFNPIDD25 pKa = 3.38TFQYY29 pKa = 10.51EE30 pKa = 4.22DD31 pKa = 3.86AEE33 pKa = 4.55GNEE36 pKa = 4.05PNDD39 pKa = 3.7RR40 pKa = 11.84KK41 pKa = 10.61SGYY44 pKa = 7.18QLKK47 pKa = 10.11LDD49 pKa = 3.86CAGIEE54 pKa = 4.08GDD56 pKa = 3.69PVEE59 pKa = 5.34SINATPAPVRR69 pKa = 11.84PPTPSPTAAATPGTSEE85 pKa = 3.95VTIAPSLEE93 pKa = 4.13AATTAAPTGMPAPSSTVSATRR114 pKa = 11.84GPFKK118 pKa = 10.67VTMAPSLEE126 pKa = 4.16ATTAAPTGMEE136 pKa = 3.6TDD138 pKa = 3.18AWLDD142 pKa = 3.49AATSAPSTVATGSPTEE158 pKa = 4.08MVMDD162 pKa = 4.52PWLAAATPAPTMRR175 pKa = 11.84TTAAPTQMATDD186 pKa = 4.57PLLQAATPAPTTQTTAGPTEE206 pKa = 4.57LVTDD210 pKa = 4.65PLLEE214 pKa = 5.03ADD216 pKa = 4.43APVPTTGTTAAPTEE230 pKa = 4.16MAMDD234 pKa = 5.05PSLEE238 pKa = 4.16AATPAPTTQTTAGPTEE254 pKa = 4.57LVTDD258 pKa = 4.65PLLEE262 pKa = 5.03ADD264 pKa = 4.43APVPTTGTTAAPTEE278 pKa = 4.16MAMDD282 pKa = 5.05PSLEE286 pKa = 4.15AATPTPSTVASAAPTKK302 pKa = 10.31MEE304 pKa = 4.5MDD306 pKa = 4.24PSLEE310 pKa = 4.11AATPTPSTVASAAPTKK326 pKa = 10.31MEE328 pKa = 4.5MDD330 pKa = 4.24PSLEE334 pKa = 4.11AATPTPSTVASAAPTKK350 pKa = 10.31MEE352 pKa = 4.5MDD354 pKa = 4.24PSLEE358 pKa = 4.11AATPTPSTVASAAPTEE374 pKa = 4.34MVMDD378 pKa = 5.1PLLGADD384 pKa = 3.91APAPTTATMAPSLEE398 pKa = 4.26AVTASPTEE406 pKa = 3.83MALDD410 pKa = 3.65KK411 pKa = 10.92SLEE414 pKa = 4.43SVTPAPTTQTTAGPTEE430 pKa = 4.57LVTDD434 pKa = 4.52PLLEE438 pKa = 4.98AGAPAPTTGTTAAPTEE454 pKa = 4.16MAMDD458 pKa = 5.05PSLEE462 pKa = 4.15AATPTPSTVASAAPTKK478 pKa = 10.25MVMDD482 pKa = 5.25PSLEE486 pKa = 4.13AATPTPSTVASVAPTEE502 pKa = 4.14MVMDD506 pKa = 5.13PSLEE510 pKa = 4.11AATPTPHH517 pKa = 7.39ADD519 pKa = 3.07TVYY522 pKa = 11.0GSVGRR527 pKa = 11.84TNADD531 pKa = 2.51GHH533 pKa = 6.08GPIAGGSHH541 pKa = 6.52VGTDD545 pKa = 3.61YY546 pKa = 11.58GNHH549 pKa = 6.79GGTDD553 pKa = 3.47TDD555 pKa = 3.35GDD557 pKa = 3.91RR558 pKa = 11.84PIASGSHH565 pKa = 6.88ADD567 pKa = 3.59TDD569 pKa = 4.28HH570 pKa = 6.97ANHH573 pKa = 7.09GGTDD577 pKa = 3.61ADD579 pKa = 3.58SDD581 pKa = 4.35GPITGGSHH589 pKa = 6.66VGTDD593 pKa = 3.61YY594 pKa = 11.57GNHH597 pKa = 6.73GGTHH601 pKa = 6.63ANGDD605 pKa = 3.8GPIASGSHH613 pKa = 6.96ADD615 pKa = 3.08TDD617 pKa = 4.09YY618 pKa = 11.95ANDD621 pKa = 3.21GRR623 pKa = 11.84TDD625 pKa = 3.66RR626 pKa = 11.84LCNGFSDD633 pKa = 3.94YY634 pKa = 11.0HH635 pKa = 7.56YY636 pKa = 11.36LLAWYY641 pKa = 9.31DD642 pKa = 3.58SSTGSWNSSTDD653 pKa = 2.6TDD655 pKa = 4.02YY656 pKa = 12.02ANDD659 pKa = 3.26GRR661 pKa = 11.84TDD663 pKa = 3.66RR664 pKa = 11.84LCNGFSDD671 pKa = 3.94YY672 pKa = 11.0HH673 pKa = 7.56YY674 pKa = 11.36LLAWYY679 pKa = 9.31DD680 pKa = 3.58SSTGSWNSSTDD691 pKa = 2.6TDD693 pKa = 4.02YY694 pKa = 12.02ANDD697 pKa = 3.26GRR699 pKa = 11.84TDD701 pKa = 3.66RR702 pKa = 11.84LCNGFSDD709 pKa = 3.76YY710 pKa = 11.0HH711 pKa = 7.65YY712 pKa = 11.35LLTWYY717 pKa = 9.99DD718 pKa = 3.22SSTGSRR724 pKa = 11.84NSSTDD729 pKa = 2.78PWNSSAVGGG738 pKa = 4.01
MM1 pKa = 7.66SLEE4 pKa = 4.19FRR6 pKa = 11.84LDD8 pKa = 3.2LCGYY12 pKa = 10.14CDD14 pKa = 3.55TYY16 pKa = 11.96NNGSFNPIDD25 pKa = 3.38TFQYY29 pKa = 10.51EE30 pKa = 4.22DD31 pKa = 3.86AEE33 pKa = 4.55GNEE36 pKa = 4.05PNDD39 pKa = 3.7RR40 pKa = 11.84KK41 pKa = 10.61SGYY44 pKa = 7.18QLKK47 pKa = 10.11LDD49 pKa = 3.86CAGIEE54 pKa = 4.08GDD56 pKa = 3.69PVEE59 pKa = 5.34SINATPAPVRR69 pKa = 11.84PPTPSPTAAATPGTSEE85 pKa = 3.95VTIAPSLEE93 pKa = 4.13AATTAAPTGMPAPSSTVSATRR114 pKa = 11.84GPFKK118 pKa = 10.67VTMAPSLEE126 pKa = 4.16ATTAAPTGMEE136 pKa = 3.6TDD138 pKa = 3.18AWLDD142 pKa = 3.49AATSAPSTVATGSPTEE158 pKa = 4.08MVMDD162 pKa = 4.52PWLAAATPAPTMRR175 pKa = 11.84TTAAPTQMATDD186 pKa = 4.57PLLQAATPAPTTQTTAGPTEE206 pKa = 4.57LVTDD210 pKa = 4.65PLLEE214 pKa = 5.03ADD216 pKa = 4.43APVPTTGTTAAPTEE230 pKa = 4.16MAMDD234 pKa = 5.05PSLEE238 pKa = 4.16AATPAPTTQTTAGPTEE254 pKa = 4.57LVTDD258 pKa = 4.65PLLEE262 pKa = 5.03ADD264 pKa = 4.43APVPTTGTTAAPTEE278 pKa = 4.16MAMDD282 pKa = 5.05PSLEE286 pKa = 4.15AATPTPSTVASAAPTKK302 pKa = 10.31MEE304 pKa = 4.5MDD306 pKa = 4.24PSLEE310 pKa = 4.11AATPTPSTVASAAPTKK326 pKa = 10.31MEE328 pKa = 4.5MDD330 pKa = 4.24PSLEE334 pKa = 4.11AATPTPSTVASAAPTKK350 pKa = 10.31MEE352 pKa = 4.5MDD354 pKa = 4.24PSLEE358 pKa = 4.11AATPTPSTVASAAPTEE374 pKa = 4.34MVMDD378 pKa = 5.1PLLGADD384 pKa = 3.91APAPTTATMAPSLEE398 pKa = 4.26AVTASPTEE406 pKa = 3.83MALDD410 pKa = 3.65KK411 pKa = 10.92SLEE414 pKa = 4.43SVTPAPTTQTTAGPTEE430 pKa = 4.57LVTDD434 pKa = 4.52PLLEE438 pKa = 4.98AGAPAPTTGTTAAPTEE454 pKa = 4.16MAMDD458 pKa = 5.05PSLEE462 pKa = 4.15AATPTPSTVASAAPTKK478 pKa = 10.25MVMDD482 pKa = 5.25PSLEE486 pKa = 4.13AATPTPSTVASVAPTEE502 pKa = 4.14MVMDD506 pKa = 5.13PSLEE510 pKa = 4.11AATPTPHH517 pKa = 7.39ADD519 pKa = 3.07TVYY522 pKa = 11.0GSVGRR527 pKa = 11.84TNADD531 pKa = 2.51GHH533 pKa = 6.08GPIAGGSHH541 pKa = 6.52VGTDD545 pKa = 3.61YY546 pKa = 11.58GNHH549 pKa = 6.79GGTDD553 pKa = 3.47TDD555 pKa = 3.35GDD557 pKa = 3.91RR558 pKa = 11.84PIASGSHH565 pKa = 6.88ADD567 pKa = 3.59TDD569 pKa = 4.28HH570 pKa = 6.97ANHH573 pKa = 7.09GGTDD577 pKa = 3.61ADD579 pKa = 3.58SDD581 pKa = 4.35GPITGGSHH589 pKa = 6.66VGTDD593 pKa = 3.61YY594 pKa = 11.57GNHH597 pKa = 6.73GGTHH601 pKa = 6.63ANGDD605 pKa = 3.8GPIASGSHH613 pKa = 6.96ADD615 pKa = 3.08TDD617 pKa = 4.09YY618 pKa = 11.95ANDD621 pKa = 3.21GRR623 pKa = 11.84TDD625 pKa = 3.66RR626 pKa = 11.84LCNGFSDD633 pKa = 3.94YY634 pKa = 11.0HH635 pKa = 7.56YY636 pKa = 11.36LLAWYY641 pKa = 9.31DD642 pKa = 3.58SSTGSWNSSTDD653 pKa = 2.6TDD655 pKa = 4.02YY656 pKa = 12.02ANDD659 pKa = 3.26GRR661 pKa = 11.84TDD663 pKa = 3.66RR664 pKa = 11.84LCNGFSDD671 pKa = 3.94YY672 pKa = 11.0HH673 pKa = 7.56YY674 pKa = 11.36LLAWYY679 pKa = 9.31DD680 pKa = 3.58SSTGSWNSSTDD691 pKa = 2.6TDD693 pKa = 4.02YY694 pKa = 12.02ANDD697 pKa = 3.26GRR699 pKa = 11.84TDD701 pKa = 3.66RR702 pKa = 11.84LCNGFSDD709 pKa = 3.76YY710 pKa = 11.0HH711 pKa = 7.65YY712 pKa = 11.35LLTWYY717 pKa = 9.99DD718 pKa = 3.22SSTGSRR724 pKa = 11.84NSSTDD729 pKa = 2.78PWNSSAVGGG738 pKa = 4.01
Molecular weight: 75.04 kDa
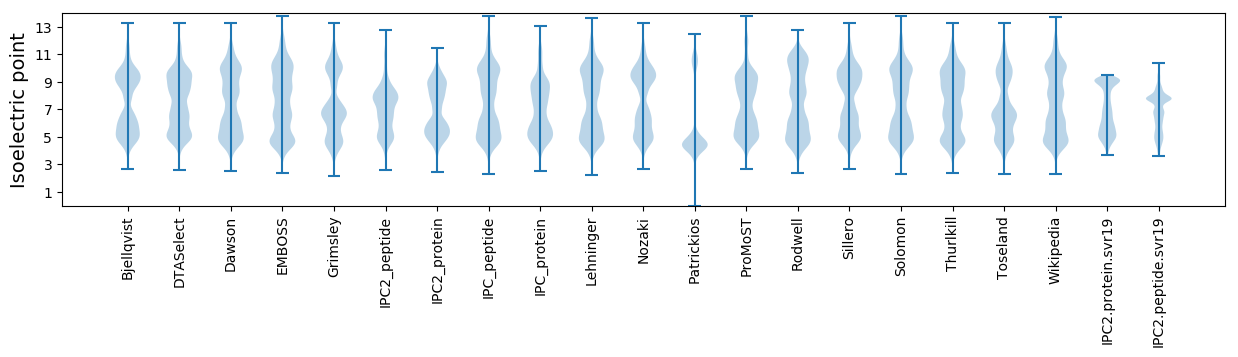
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6H5KJ57|A0A6H5KJ57_9PHAE Uncharacterized protein OS=Ectocarpus sp. CCAP 1310/34 OX=867726 GN=ESUBFT_41_3 PE=4 SV=1
MM1 pKa = 7.47SSSSTTVHH9 pKa = 5.82SRR11 pKa = 11.84RR12 pKa = 11.84PLPPPLSLSSSRR24 pKa = 11.84RR25 pKa = 11.84RR26 pKa = 11.84STSSRR31 pKa = 11.84RR32 pKa = 11.84FPPAPLRR39 pKa = 11.84GTWRR43 pKa = 11.84VPSRR47 pKa = 11.84QVV49 pKa = 2.6
MM1 pKa = 7.47SSSSTTVHH9 pKa = 5.82SRR11 pKa = 11.84RR12 pKa = 11.84PLPPPLSLSSSRR24 pKa = 11.84RR25 pKa = 11.84RR26 pKa = 11.84STSSRR31 pKa = 11.84RR32 pKa = 11.84FPPAPLRR39 pKa = 11.84GTWRR43 pKa = 11.84VPSRR47 pKa = 11.84QVV49 pKa = 2.6
Molecular weight: 5.5 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
11448718 |
20 |
11060 |
452.0 |
48.21 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.134 ± 0.033 | 1.62 ± 0.007 |
5.512 ± 0.014 | 6.582 ± 0.018 |
2.935 ± 0.011 | 10.056 ± 0.028 |
2.105 ± 0.007 | 2.946 ± 0.01 |
4.35 ± 0.018 | 7.88 ± 0.019 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.156 ± 0.007 | 2.697 ± 0.01 |
5.486 ± 0.019 | 3.439 ± 0.012 |
7.305 ± 0.016 | 7.844 ± 0.022 |
5.331 ± 0.014 | 6.82 ± 0.015 |
1.299 ± 0.006 | 1.828 ± 0.009 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
