
Gloeothece citriformis (strain PCC 7424) (Cyanothece sp. (strain PCC 7424))
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Cyanobacteria/Melainabacteria group; Cyanobacteria; Oscillatoriophycideae; Chroococcales; Aphanothecaceae; Gloeothece; Gloeothece citriformis
Average proteome isoelectric point is 6.24
Get precalculated fractions of proteins
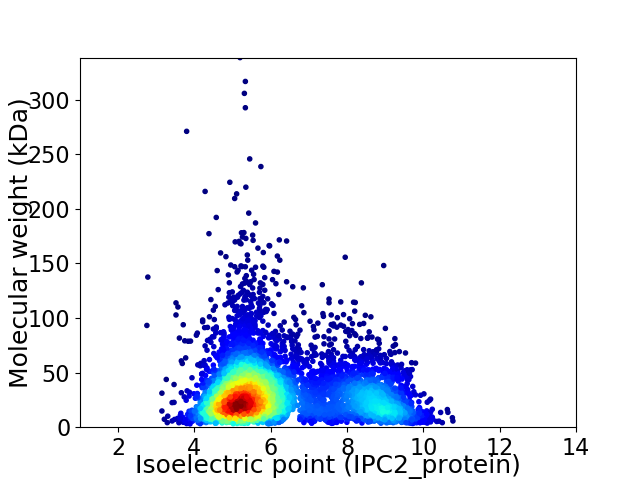
Virtual 2D-PAGE plot for 5656 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
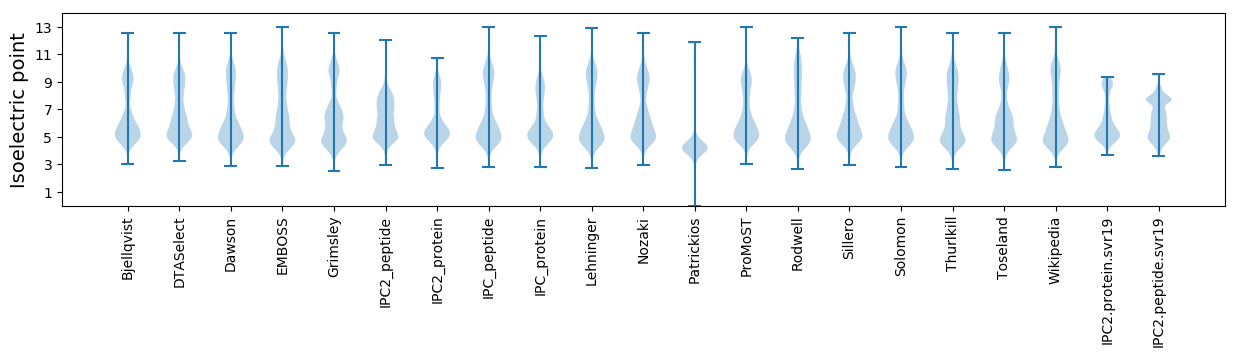
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|B7KF08|PYRG_GLOC7 CTP synthase OS=Gloeothece citriformis (strain PCC 7424) OX=65393 GN=pyrG PE=3 SV=1
MM1 pKa = 7.79AVFTVTNLLDD11 pKa = 3.86NGEE14 pKa = 4.12GSLRR18 pKa = 11.84QAIANANSLDD28 pKa = 3.79GADD31 pKa = 3.79TIEE34 pKa = 4.25FASNLSGGTISLTSGEE50 pKa = 4.22LSITDD55 pKa = 3.73SLTLEE60 pKa = 4.5GLGSDD65 pKa = 4.66LLTISGSDD73 pKa = 3.36LSRR76 pKa = 11.84IFNINDD82 pKa = 2.86QTEE85 pKa = 4.66YY86 pKa = 11.24YY87 pKa = 10.12SYY89 pKa = 11.33NSNILDD95 pKa = 3.7VVISGLTLTNGFVSEE110 pKa = 4.21NTQSFNAPAASGGAIYY126 pKa = 10.36NYY128 pKa = 10.43DD129 pKa = 3.85EE130 pKa = 4.65NLTLIDD136 pKa = 3.77TVITGNTAAGDD147 pKa = 3.49PFAYY151 pKa = 10.44YY152 pKa = 10.57EE153 pKa = 4.01NAEE156 pKa = 4.11ALGGGIFNYY165 pKa = 10.35SGTVTVIDD173 pKa = 4.21SVISSNTAQGGTVLLDD189 pKa = 4.46FIRR192 pKa = 11.84EE193 pKa = 4.14GGFVGSGGGIYY204 pKa = 10.3SRR206 pKa = 11.84TGNVTVINSTISDD219 pKa = 3.61NTAKK223 pKa = 10.03TGRR226 pKa = 11.84GGDD229 pKa = 3.51FVFTYY234 pKa = 9.34EE235 pKa = 4.04SHH237 pKa = 6.55GGGIAGGFNSTITITNSTISNNTVVGANTTNIYY270 pKa = 10.36SRR272 pKa = 11.84EE273 pKa = 4.14GGDD276 pKa = 3.33ARR278 pKa = 11.84GGGISGDD285 pKa = 3.24NVIVNNSSITGNTVLGGNGTGSFYY309 pKa = 11.09VYY311 pKa = 10.05YY312 pKa = 9.78GYY314 pKa = 10.91SGGYY318 pKa = 9.86GGDD321 pKa = 3.26ARR323 pKa = 11.84GGGISGNNVTVSNSTISGNIAQGGNGGIGGKK354 pKa = 10.5ANVFRR359 pKa = 11.84GFFYY363 pKa = 9.49GTNGGRR369 pKa = 11.84GGNATGGGISSANLTMSNSTISGNTAQGGNGANGGQGADD408 pKa = 2.74TATINEE414 pKa = 4.06YY415 pKa = 10.4FGYY418 pKa = 9.8SGYY421 pKa = 11.18YY422 pKa = 10.05YY423 pKa = 10.85YY424 pKa = 10.9FAGNGGNGGNGGNSVGGGVAVSNNALIANSTITNNTLDD462 pKa = 3.81SGTGGVGGSGGLGNEE477 pKa = 4.58SLNIPNGIDD486 pKa = 3.54GTDD489 pKa = 3.3GTIGNEE495 pKa = 3.57IGGGLSANGTVTSTIIAGNTDD516 pKa = 3.37NQDD519 pKa = 3.35LAGTFTSGGNNLIGNGDD536 pKa = 3.85GATGFIDD543 pKa = 4.82GVNDD547 pKa = 3.78DD548 pKa = 4.38QVGTTINPIDD558 pKa = 4.47PLLSPLQDD566 pKa = 3.32NGGLTLTHH574 pKa = 6.64TLLPGSPAIDD584 pKa = 3.69AGSNPSTLISDD595 pKa = 3.33QRR597 pKa = 11.84GAPRR601 pKa = 11.84VAGDD605 pKa = 3.63GADD608 pKa = 3.15IGALEE613 pKa = 4.54LADD616 pKa = 3.85EE617 pKa = 4.6TLLGDD622 pKa = 3.49QSSNSFTSGTGNDD635 pKa = 3.69FLEE638 pKa = 5.31GGDD641 pKa = 5.02GNDD644 pKa = 3.98ALNGSSGNDD653 pKa = 3.62GLRR656 pKa = 11.84GGAGSDD662 pKa = 3.39RR663 pKa = 11.84LIGSDD668 pKa = 4.96GNDD671 pKa = 3.57LLIGGLDD678 pKa = 3.52RR679 pKa = 11.84DD680 pKa = 3.69ILAGGEE686 pKa = 4.13GADD689 pKa = 3.29VFRR692 pKa = 11.84FEE694 pKa = 4.71SLADD698 pKa = 3.74SSLLSPDD705 pKa = 4.32RR706 pKa = 11.84IRR708 pKa = 11.84DD709 pKa = 3.73FNQTEE714 pKa = 4.1GDD716 pKa = 3.78RR717 pKa = 11.84FQLLPTNPSTLWNAGDD733 pKa = 3.61VTGDD737 pKa = 3.48NLRR740 pKa = 11.84VALEE744 pKa = 3.66AAYY747 pKa = 10.46ADD749 pKa = 4.38SNQSAGGSQPLGLNEE764 pKa = 3.78AVLFNFGSRR773 pKa = 11.84SYY775 pKa = 11.43LSINNTISGFNPEE788 pKa = 3.49QDD790 pKa = 3.2LVVEE794 pKa = 4.19VSGIVLASGDD804 pKa = 3.82EE805 pKa = 4.22NAGVLSVSDD814 pKa = 3.81YY815 pKa = 11.13FVV817 pKa = 3.0
MM1 pKa = 7.79AVFTVTNLLDD11 pKa = 3.86NGEE14 pKa = 4.12GSLRR18 pKa = 11.84QAIANANSLDD28 pKa = 3.79GADD31 pKa = 3.79TIEE34 pKa = 4.25FASNLSGGTISLTSGEE50 pKa = 4.22LSITDD55 pKa = 3.73SLTLEE60 pKa = 4.5GLGSDD65 pKa = 4.66LLTISGSDD73 pKa = 3.36LSRR76 pKa = 11.84IFNINDD82 pKa = 2.86QTEE85 pKa = 4.66YY86 pKa = 11.24YY87 pKa = 10.12SYY89 pKa = 11.33NSNILDD95 pKa = 3.7VVISGLTLTNGFVSEE110 pKa = 4.21NTQSFNAPAASGGAIYY126 pKa = 10.36NYY128 pKa = 10.43DD129 pKa = 3.85EE130 pKa = 4.65NLTLIDD136 pKa = 3.77TVITGNTAAGDD147 pKa = 3.49PFAYY151 pKa = 10.44YY152 pKa = 10.57EE153 pKa = 4.01NAEE156 pKa = 4.11ALGGGIFNYY165 pKa = 10.35SGTVTVIDD173 pKa = 4.21SVISSNTAQGGTVLLDD189 pKa = 4.46FIRR192 pKa = 11.84EE193 pKa = 4.14GGFVGSGGGIYY204 pKa = 10.3SRR206 pKa = 11.84TGNVTVINSTISDD219 pKa = 3.61NTAKK223 pKa = 10.03TGRR226 pKa = 11.84GGDD229 pKa = 3.51FVFTYY234 pKa = 9.34EE235 pKa = 4.04SHH237 pKa = 6.55GGGIAGGFNSTITITNSTISNNTVVGANTTNIYY270 pKa = 10.36SRR272 pKa = 11.84EE273 pKa = 4.14GGDD276 pKa = 3.33ARR278 pKa = 11.84GGGISGDD285 pKa = 3.24NVIVNNSSITGNTVLGGNGTGSFYY309 pKa = 11.09VYY311 pKa = 10.05YY312 pKa = 9.78GYY314 pKa = 10.91SGGYY318 pKa = 9.86GGDD321 pKa = 3.26ARR323 pKa = 11.84GGGISGNNVTVSNSTISGNIAQGGNGGIGGKK354 pKa = 10.5ANVFRR359 pKa = 11.84GFFYY363 pKa = 9.49GTNGGRR369 pKa = 11.84GGNATGGGISSANLTMSNSTISGNTAQGGNGANGGQGADD408 pKa = 2.74TATINEE414 pKa = 4.06YY415 pKa = 10.4FGYY418 pKa = 9.8SGYY421 pKa = 11.18YY422 pKa = 10.05YY423 pKa = 10.85YY424 pKa = 10.9FAGNGGNGGNGGNSVGGGVAVSNNALIANSTITNNTLDD462 pKa = 3.81SGTGGVGGSGGLGNEE477 pKa = 4.58SLNIPNGIDD486 pKa = 3.54GTDD489 pKa = 3.3GTIGNEE495 pKa = 3.57IGGGLSANGTVTSTIIAGNTDD516 pKa = 3.37NQDD519 pKa = 3.35LAGTFTSGGNNLIGNGDD536 pKa = 3.85GATGFIDD543 pKa = 4.82GVNDD547 pKa = 3.78DD548 pKa = 4.38QVGTTINPIDD558 pKa = 4.47PLLSPLQDD566 pKa = 3.32NGGLTLTHH574 pKa = 6.64TLLPGSPAIDD584 pKa = 3.69AGSNPSTLISDD595 pKa = 3.33QRR597 pKa = 11.84GAPRR601 pKa = 11.84VAGDD605 pKa = 3.63GADD608 pKa = 3.15IGALEE613 pKa = 4.54LADD616 pKa = 3.85EE617 pKa = 4.6TLLGDD622 pKa = 3.49QSSNSFTSGTGNDD635 pKa = 3.69FLEE638 pKa = 5.31GGDD641 pKa = 5.02GNDD644 pKa = 3.98ALNGSSGNDD653 pKa = 3.62GLRR656 pKa = 11.84GGAGSDD662 pKa = 3.39RR663 pKa = 11.84LIGSDD668 pKa = 4.96GNDD671 pKa = 3.57LLIGGLDD678 pKa = 3.52RR679 pKa = 11.84DD680 pKa = 3.69ILAGGEE686 pKa = 4.13GADD689 pKa = 3.29VFRR692 pKa = 11.84FEE694 pKa = 4.71SLADD698 pKa = 3.74SSLLSPDD705 pKa = 4.32RR706 pKa = 11.84IRR708 pKa = 11.84DD709 pKa = 3.73FNQTEE714 pKa = 4.1GDD716 pKa = 3.78RR717 pKa = 11.84FQLLPTNPSTLWNAGDD733 pKa = 3.61VTGDD737 pKa = 3.48NLRR740 pKa = 11.84VALEE744 pKa = 3.66AAYY747 pKa = 10.46ADD749 pKa = 4.38SNQSAGGSQPLGLNEE764 pKa = 3.78AVLFNFGSRR773 pKa = 11.84SYY775 pKa = 11.43LSINNTISGFNPEE788 pKa = 3.49QDD790 pKa = 3.2LVVEE794 pKa = 4.19VSGIVLASGDD804 pKa = 3.82EE805 pKa = 4.22NAGVLSVSDD814 pKa = 3.81YY815 pKa = 11.13FVV817 pKa = 3.0
Molecular weight: 81.66 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|B7KHH1|B7KHH1_GLOC7 Extracellular solute-binding protein family 1 OS=Gloeothece citriformis (strain PCC 7424) OX=65393 GN=PCC7424_2244 PE=4 SV=1
MM1 pKa = 7.29TRR3 pKa = 11.84RR4 pKa = 11.84TLEE7 pKa = 3.71GTSRR11 pKa = 11.84KK12 pKa = 9.03RR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84VSGFRR20 pKa = 11.84TRR22 pKa = 11.84MRR24 pKa = 11.84TINGRR29 pKa = 11.84KK30 pKa = 8.8VIQARR35 pKa = 11.84RR36 pKa = 11.84QKK38 pKa = 10.3GRR40 pKa = 11.84HH41 pKa = 5.42KK42 pKa = 10.85LSVSEE47 pKa = 4.34GG48 pKa = 3.44
MM1 pKa = 7.29TRR3 pKa = 11.84RR4 pKa = 11.84TLEE7 pKa = 3.71GTSRR11 pKa = 11.84KK12 pKa = 9.03RR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84VSGFRR20 pKa = 11.84TRR22 pKa = 11.84MRR24 pKa = 11.84TINGRR29 pKa = 11.84KK30 pKa = 8.8VIQARR35 pKa = 11.84RR36 pKa = 11.84QKK38 pKa = 10.3GRR40 pKa = 11.84HH41 pKa = 5.42KK42 pKa = 10.85LSVSEE47 pKa = 4.34GG48 pKa = 3.44
Molecular weight: 5.7 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1754939 |
29 |
2997 |
310.3 |
34.9 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.748 ± 0.035 | 1.006 ± 0.011 |
4.856 ± 0.025 | 6.686 ± 0.036 |
4.084 ± 0.024 | 6.422 ± 0.04 |
1.844 ± 0.016 | 7.396 ± 0.029 |
5.47 ± 0.033 | 11.219 ± 0.042 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.804 ± 0.015 | 4.687 ± 0.029 |
4.741 ± 0.029 | 5.476 ± 0.034 |
4.742 ± 0.025 | 6.435 ± 0.028 |
5.586 ± 0.027 | 6.035 ± 0.028 |
1.476 ± 0.015 | 3.287 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
