
Marine Group I thaumarchaeote SCGC AAA799-B03
Taxonomy: cellular organisms; Archaea; TACK group; Thaumarchaeota; Thaumarchaeota incertae sedis; Marine Group I
Average proteome isoelectric point is 6.45
Get precalculated fractions of proteins
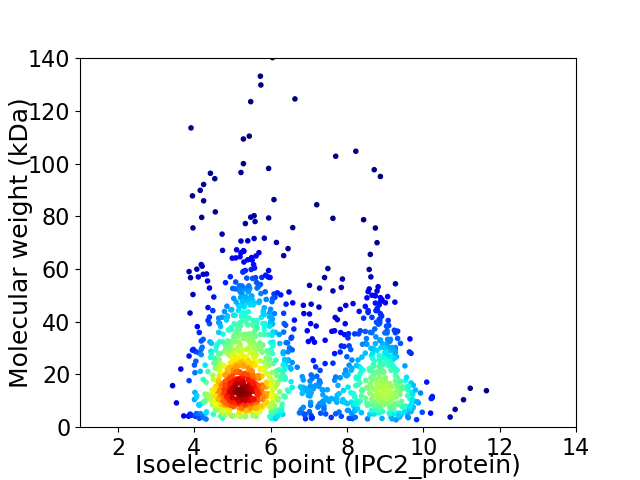
Virtual 2D-PAGE plot for 1339 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A087S7R3|A0A087S7R3_9ARCH Phosphate transport system permease protein PstA OS=Marine Group I thaumarchaeote SCGC AAA799-B03 OX=1502289 GN=pstA PE=3 SV=1
MM1 pKa = 7.04KK2 pKa = 9.13TLPVIVTTLSVILVSSTLTPSFALGEE28 pKa = 3.94YY29 pKa = 10.06DD30 pKa = 5.24YY31 pKa = 11.46LSEE34 pKa = 3.78WGEE37 pKa = 3.87FGIYY41 pKa = 9.42TPGFFSYY48 pKa = 10.58PEE50 pKa = 4.09YY51 pKa = 10.58IAVDD55 pKa = 3.67EE56 pKa = 5.45DD57 pKa = 4.05GNSYY61 pKa = 10.67ISDD64 pKa = 4.06LGNKK68 pKa = 9.7RR69 pKa = 11.84IQKK72 pKa = 9.96FSSDD76 pKa = 3.35GQYY79 pKa = 9.87ISHH82 pKa = 6.99WGEE85 pKa = 3.75SGKK88 pKa = 9.89QAGQFHH94 pKa = 6.84YY95 pKa = 9.96PSGITISGDD104 pKa = 3.13HH105 pKa = 6.74VYY107 pKa = 10.74VADD110 pKa = 3.86QALHH114 pKa = 7.27RR115 pKa = 11.84IQQFTLDD122 pKa = 4.22GEE124 pKa = 5.18FVSQWGSKK132 pKa = 9.85GIHH135 pKa = 5.78EE136 pKa = 5.08GEE138 pKa = 3.9FKK140 pKa = 10.82YY141 pKa = 10.33PYY143 pKa = 10.24GITSDD148 pKa = 3.86SEE150 pKa = 3.98HH151 pKa = 7.75VYY153 pKa = 10.87VVDD156 pKa = 3.9TGNQRR161 pKa = 11.84IQKK164 pKa = 8.02FTLDD168 pKa = 3.65GEE170 pKa = 4.79FVLSFGTSGMGPGQFLTATGIDD192 pKa = 3.55VEE194 pKa = 4.56DD195 pKa = 4.26DD196 pKa = 3.56VVYY199 pKa = 9.64VTDD202 pKa = 3.59KK203 pKa = 11.3GNRR206 pKa = 11.84KK207 pKa = 8.94IDD209 pKa = 3.58TFDD212 pKa = 3.48SDD214 pKa = 4.12GNFIKK219 pKa = 10.4SYY221 pKa = 9.65PFRR224 pKa = 11.84GLNYY228 pKa = 8.82VFAPEE233 pKa = 5.18GIAVSPEE240 pKa = 3.46GKK242 pKa = 10.36VFVTNSDD249 pKa = 3.71NNKK252 pKa = 9.41VLYY255 pKa = 10.6LGLDD259 pKa = 3.51GDD261 pKa = 4.3LSLDD265 pKa = 3.2IFEE268 pKa = 5.31KK269 pKa = 10.78NGPLKK274 pKa = 10.12HH275 pKa = 6.04TFTSLTDD282 pKa = 3.34VAIGSNGEE290 pKa = 4.11LLVVDD295 pKa = 4.72SAAHH299 pKa = 6.39KK300 pKa = 9.9IKK302 pKa = 10.84SFEE305 pKa = 4.13TPFYY309 pKa = 10.75SEE311 pKa = 5.23PIVKK315 pKa = 9.59EE316 pKa = 3.62QVEE319 pKa = 4.49IVAPEE324 pKa = 3.92VEE326 pKa = 4.28EE327 pKa = 5.66DD328 pKa = 3.93YY329 pKa = 11.58SFDD332 pKa = 5.83DD333 pKa = 3.61IDD335 pKa = 4.11PVIMAPSDD343 pKa = 3.32IKK345 pKa = 11.44LEE347 pKa = 4.17ATDD350 pKa = 4.86LFTPVLIGEE359 pKa = 4.66AVAEE363 pKa = 4.51DD364 pKa = 3.83LHH366 pKa = 8.53SGIKK370 pKa = 10.37AILNNAPEE378 pKa = 4.35AFSLGVSKK386 pKa = 8.65VTWIAFDD393 pKa = 3.71NAGNTAEE400 pKa = 4.94DD401 pKa = 3.89YY402 pKa = 11.59QNITVYY408 pKa = 11.08ACGNVYY414 pKa = 10.65SDD416 pKa = 3.76YY417 pKa = 11.77NMIVGTDD424 pKa = 3.65EE425 pKa = 4.84NDD427 pKa = 3.31VLQGTSGDD435 pKa = 3.78DD436 pKa = 3.96LIFGLEE442 pKa = 4.07GNDD445 pKa = 3.63IISGLEE451 pKa = 3.89GNDD454 pKa = 3.96CIFGGEE460 pKa = 4.19GDD462 pKa = 4.02DD463 pKa = 4.05VVYY466 pKa = 11.08GNDD469 pKa = 4.05GYY471 pKa = 10.58DD472 pKa = 3.41TISGNGGHH480 pKa = 7.72DD481 pKa = 3.2ILKK484 pKa = 9.97GDD486 pKa = 3.61SGFDD490 pKa = 3.45VIYY493 pKa = 10.19GGSGSDD499 pKa = 3.48VLDD502 pKa = 4.12GGSEE506 pKa = 3.96NDD508 pKa = 3.21NCYY511 pKa = 10.86DD512 pKa = 3.71SQDD515 pKa = 4.61SIILNCNEE523 pKa = 3.66
MM1 pKa = 7.04KK2 pKa = 9.13TLPVIVTTLSVILVSSTLTPSFALGEE28 pKa = 3.94YY29 pKa = 10.06DD30 pKa = 5.24YY31 pKa = 11.46LSEE34 pKa = 3.78WGEE37 pKa = 3.87FGIYY41 pKa = 9.42TPGFFSYY48 pKa = 10.58PEE50 pKa = 4.09YY51 pKa = 10.58IAVDD55 pKa = 3.67EE56 pKa = 5.45DD57 pKa = 4.05GNSYY61 pKa = 10.67ISDD64 pKa = 4.06LGNKK68 pKa = 9.7RR69 pKa = 11.84IQKK72 pKa = 9.96FSSDD76 pKa = 3.35GQYY79 pKa = 9.87ISHH82 pKa = 6.99WGEE85 pKa = 3.75SGKK88 pKa = 9.89QAGQFHH94 pKa = 6.84YY95 pKa = 9.96PSGITISGDD104 pKa = 3.13HH105 pKa = 6.74VYY107 pKa = 10.74VADD110 pKa = 3.86QALHH114 pKa = 7.27RR115 pKa = 11.84IQQFTLDD122 pKa = 4.22GEE124 pKa = 5.18FVSQWGSKK132 pKa = 9.85GIHH135 pKa = 5.78EE136 pKa = 5.08GEE138 pKa = 3.9FKK140 pKa = 10.82YY141 pKa = 10.33PYY143 pKa = 10.24GITSDD148 pKa = 3.86SEE150 pKa = 3.98HH151 pKa = 7.75VYY153 pKa = 10.87VVDD156 pKa = 3.9TGNQRR161 pKa = 11.84IQKK164 pKa = 8.02FTLDD168 pKa = 3.65GEE170 pKa = 4.79FVLSFGTSGMGPGQFLTATGIDD192 pKa = 3.55VEE194 pKa = 4.56DD195 pKa = 4.26DD196 pKa = 3.56VVYY199 pKa = 9.64VTDD202 pKa = 3.59KK203 pKa = 11.3GNRR206 pKa = 11.84KK207 pKa = 8.94IDD209 pKa = 3.58TFDD212 pKa = 3.48SDD214 pKa = 4.12GNFIKK219 pKa = 10.4SYY221 pKa = 9.65PFRR224 pKa = 11.84GLNYY228 pKa = 8.82VFAPEE233 pKa = 5.18GIAVSPEE240 pKa = 3.46GKK242 pKa = 10.36VFVTNSDD249 pKa = 3.71NNKK252 pKa = 9.41VLYY255 pKa = 10.6LGLDD259 pKa = 3.51GDD261 pKa = 4.3LSLDD265 pKa = 3.2IFEE268 pKa = 5.31KK269 pKa = 10.78NGPLKK274 pKa = 10.12HH275 pKa = 6.04TFTSLTDD282 pKa = 3.34VAIGSNGEE290 pKa = 4.11LLVVDD295 pKa = 4.72SAAHH299 pKa = 6.39KK300 pKa = 9.9IKK302 pKa = 10.84SFEE305 pKa = 4.13TPFYY309 pKa = 10.75SEE311 pKa = 5.23PIVKK315 pKa = 9.59EE316 pKa = 3.62QVEE319 pKa = 4.49IVAPEE324 pKa = 3.92VEE326 pKa = 4.28EE327 pKa = 5.66DD328 pKa = 3.93YY329 pKa = 11.58SFDD332 pKa = 5.83DD333 pKa = 3.61IDD335 pKa = 4.11PVIMAPSDD343 pKa = 3.32IKK345 pKa = 11.44LEE347 pKa = 4.17ATDD350 pKa = 4.86LFTPVLIGEE359 pKa = 4.66AVAEE363 pKa = 4.51DD364 pKa = 3.83LHH366 pKa = 8.53SGIKK370 pKa = 10.37AILNNAPEE378 pKa = 4.35AFSLGVSKK386 pKa = 8.65VTWIAFDD393 pKa = 3.71NAGNTAEE400 pKa = 4.94DD401 pKa = 3.89YY402 pKa = 11.59QNITVYY408 pKa = 11.08ACGNVYY414 pKa = 10.65SDD416 pKa = 3.76YY417 pKa = 11.77NMIVGTDD424 pKa = 3.65EE425 pKa = 4.84NDD427 pKa = 3.31VLQGTSGDD435 pKa = 3.78DD436 pKa = 3.96LIFGLEE442 pKa = 4.07GNDD445 pKa = 3.63IISGLEE451 pKa = 3.89GNDD454 pKa = 3.96CIFGGEE460 pKa = 4.19GDD462 pKa = 4.02DD463 pKa = 4.05VVYY466 pKa = 11.08GNDD469 pKa = 4.05GYY471 pKa = 10.58DD472 pKa = 3.41TISGNGGHH480 pKa = 7.72DD481 pKa = 3.2ILKK484 pKa = 9.97GDD486 pKa = 3.61SGFDD490 pKa = 3.45VIYY493 pKa = 10.19GGSGSDD499 pKa = 3.48VLDD502 pKa = 4.12GGSEE506 pKa = 3.96NDD508 pKa = 3.21NCYY511 pKa = 10.86DD512 pKa = 3.71SQDD515 pKa = 4.61SIILNCNEE523 pKa = 3.66
Molecular weight: 56.7 kDa
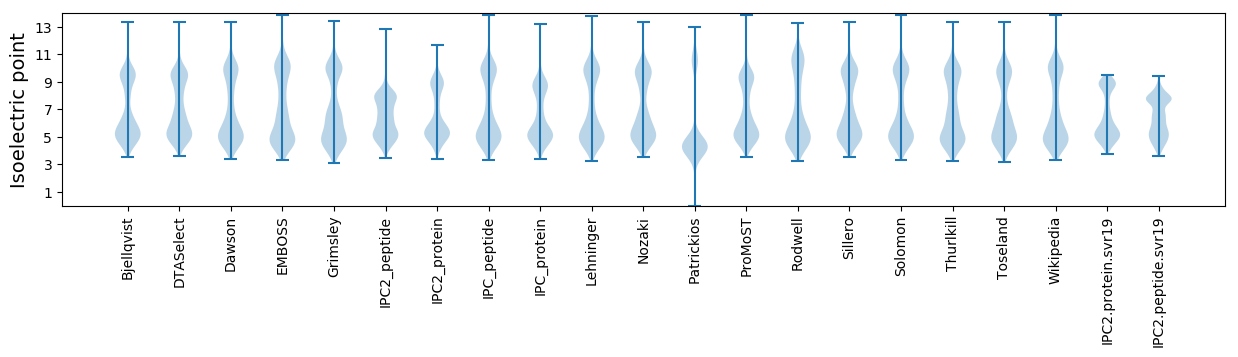
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A087S8E1|A0A087S8E1_9ARCH Uncharacterized protein OS=Marine Group I thaumarchaeote SCGC AAA799-B03 OX=1502289 GN=AAA799B03_00442 PE=4 SV=1
MM1 pKa = 7.42AMSRR5 pKa = 11.84AKK7 pKa = 10.31RR8 pKa = 11.84AEE10 pKa = 3.95AAKK13 pKa = 8.98KK14 pKa = 6.95TARR17 pKa = 11.84TRR19 pKa = 11.84KK20 pKa = 9.58RR21 pKa = 11.84NAAKK25 pKa = 10.3KK26 pKa = 9.8AAEE29 pKa = 4.03AAKK32 pKa = 10.16KK33 pKa = 9.77AAARR37 pKa = 11.84KK38 pKa = 9.21RR39 pKa = 11.84KK40 pKa = 9.17AARR43 pKa = 11.84ARR45 pKa = 11.84NAAKK49 pKa = 10.21KK50 pKa = 9.55AAPKK54 pKa = 10.27RR55 pKa = 11.84RR56 pKa = 11.84TAAKK60 pKa = 9.88RR61 pKa = 11.84KK62 pKa = 8.47AAPKK66 pKa = 10.03RR67 pKa = 11.84RR68 pKa = 11.84TAAKK72 pKa = 9.88RR73 pKa = 11.84KK74 pKa = 8.72AAPKK78 pKa = 10.04RR79 pKa = 11.84KK80 pKa = 7.37TAAKK84 pKa = 9.84RR85 pKa = 11.84KK86 pKa = 8.7AAPKK90 pKa = 10.0RR91 pKa = 11.84KK92 pKa = 9.28AATKK96 pKa = 10.14RR97 pKa = 11.84RR98 pKa = 11.84AAPKK102 pKa = 9.97RR103 pKa = 11.84KK104 pKa = 7.39TAAKK108 pKa = 9.84RR109 pKa = 11.84KK110 pKa = 8.7AAPKK114 pKa = 10.0RR115 pKa = 11.84KK116 pKa = 9.28AATKK120 pKa = 10.14RR121 pKa = 11.84RR122 pKa = 11.84AAPKK126 pKa = 9.76RR127 pKa = 11.84KK128 pKa = 8.49AAKK131 pKa = 9.71RR132 pKa = 11.84RR133 pKa = 11.84RR134 pKa = 3.63
MM1 pKa = 7.42AMSRR5 pKa = 11.84AKK7 pKa = 10.31RR8 pKa = 11.84AEE10 pKa = 3.95AAKK13 pKa = 8.98KK14 pKa = 6.95TARR17 pKa = 11.84TRR19 pKa = 11.84KK20 pKa = 9.58RR21 pKa = 11.84NAAKK25 pKa = 10.3KK26 pKa = 9.8AAEE29 pKa = 4.03AAKK32 pKa = 10.16KK33 pKa = 9.77AAARR37 pKa = 11.84KK38 pKa = 9.21RR39 pKa = 11.84KK40 pKa = 9.17AARR43 pKa = 11.84ARR45 pKa = 11.84NAAKK49 pKa = 10.21KK50 pKa = 9.55AAPKK54 pKa = 10.27RR55 pKa = 11.84RR56 pKa = 11.84TAAKK60 pKa = 9.88RR61 pKa = 11.84KK62 pKa = 8.47AAPKK66 pKa = 10.03RR67 pKa = 11.84RR68 pKa = 11.84TAAKK72 pKa = 9.88RR73 pKa = 11.84KK74 pKa = 8.72AAPKK78 pKa = 10.04RR79 pKa = 11.84KK80 pKa = 7.37TAAKK84 pKa = 9.84RR85 pKa = 11.84KK86 pKa = 8.7AAPKK90 pKa = 10.0RR91 pKa = 11.84KK92 pKa = 9.28AATKK96 pKa = 10.14RR97 pKa = 11.84RR98 pKa = 11.84AAPKK102 pKa = 9.97RR103 pKa = 11.84KK104 pKa = 7.39TAAKK108 pKa = 9.84RR109 pKa = 11.84KK110 pKa = 8.7AAPKK114 pKa = 10.0RR115 pKa = 11.84KK116 pKa = 9.28AATKK120 pKa = 10.14RR121 pKa = 11.84RR122 pKa = 11.84AAPKK126 pKa = 9.76RR127 pKa = 11.84KK128 pKa = 8.49AAKK131 pKa = 9.71RR132 pKa = 11.84RR133 pKa = 11.84RR134 pKa = 3.63
Molecular weight: 14.77 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
297946 |
23 |
1263 |
222.5 |
25.01 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.901 ± 0.082 | 1.075 ± 0.033 |
5.84 ± 0.068 | 7.408 ± 0.081 |
4.438 ± 0.061 | 6.407 ± 0.083 |
1.849 ± 0.031 | 8.676 ± 0.059 |
8.773 ± 0.091 | 8.486 ± 0.067 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.639 ± 0.041 | 4.978 ± 0.071 |
3.73 ± 0.051 | 3.231 ± 0.042 |
3.599 ± 0.05 | 7.002 ± 0.063 |
5.462 ± 0.05 | 6.443 ± 0.053 |
0.909 ± 0.029 | 3.14 ± 0.044 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
