
Grass carp reovirus
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Resentoviricetes; Reovirales; Reoviridae; Spinareovirinae; Aquareovirus; Aquareovirus C
Average proteome isoelectric point is 6.21
Get precalculated fractions of proteins
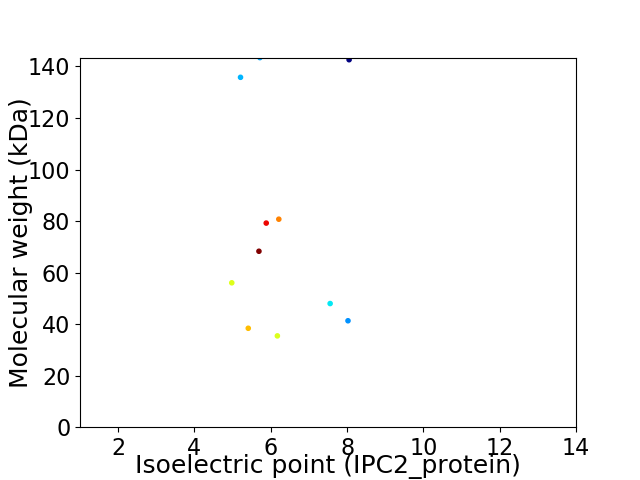
Virtual 2D-PAGE plot for 11 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
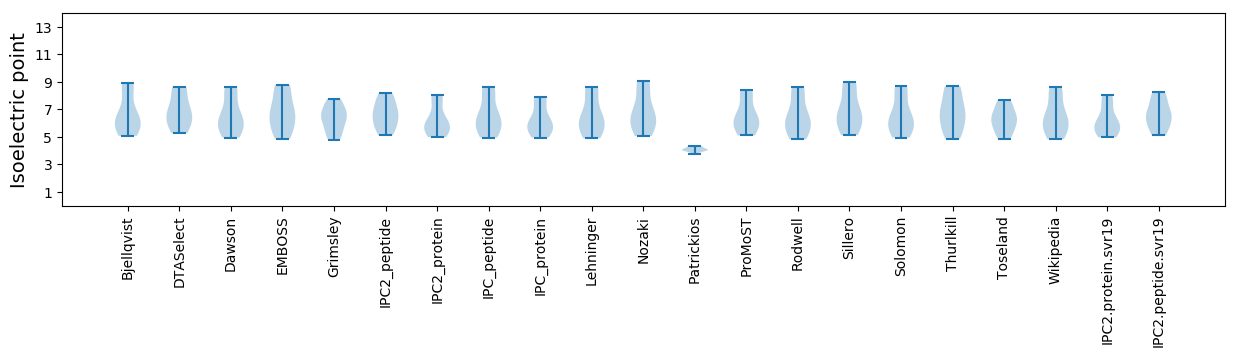
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|E7DDK9|E7DDK9_9REOV Non-structural protein OS=Grass carp reovirus OX=128987 GN=NS2 PE=4 SV=1
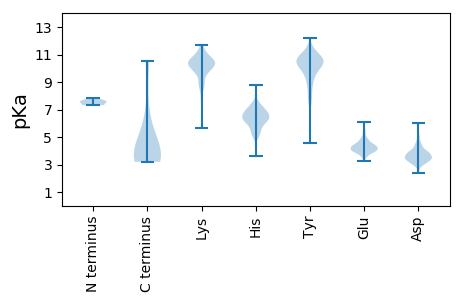
MM1 pKa = 7.37ATRR4 pKa = 11.84DD5 pKa = 3.35SRR7 pKa = 11.84MTTEE11 pKa = 4.56HH12 pKa = 6.66VIALILAYY20 pKa = 9.68STTLDD25 pKa = 3.19EE26 pKa = 4.9AKK28 pKa = 10.02IHH30 pKa = 6.25EE31 pKa = 5.49EE32 pKa = 3.53ITNLQSAVEE41 pKa = 3.95QLTVRR46 pKa = 11.84LTEE49 pKa = 3.86LTNAVTSLDD58 pKa = 3.4ARR60 pKa = 11.84LTQVEE65 pKa = 4.26QQASKK70 pKa = 11.08LDD72 pKa = 3.85DD73 pKa = 4.67KK74 pKa = 11.4IGILSNRR81 pKa = 11.84IGANTASINFLAGIVDD97 pKa = 4.06ATRR100 pKa = 11.84IDD102 pKa = 4.15AVDD105 pKa = 3.7APLYY109 pKa = 10.36IYY111 pKa = 10.38IVDD114 pKa = 3.74DD115 pKa = 3.55QRR117 pKa = 11.84RR118 pKa = 11.84LAIATGDD125 pKa = 3.62GLFVKK130 pKa = 10.19DD131 pKa = 4.43RR132 pKa = 11.84KK133 pKa = 10.75LNGYY137 pKa = 7.27DD138 pKa = 3.29VRR140 pKa = 11.84SFPPIAVAKK149 pKa = 10.34YY150 pKa = 10.17NDD152 pKa = 3.58ILSFSLSSAPPLDD165 pKa = 3.55IVDD168 pKa = 4.01GKK170 pKa = 10.89LAVSTTSRR178 pKa = 11.84LFITSGKK185 pKa = 10.06LDD187 pKa = 3.55TNSYY191 pKa = 9.64VGSSSVDD198 pKa = 2.95ISGAAAEE205 pKa = 4.42KK206 pKa = 8.81TVSVRR211 pKa = 11.84TINPIIAGQDD221 pKa = 3.54GLSLSLSGVLEE232 pKa = 4.3VGSGLLLSGRR242 pKa = 11.84SWPLTLDD249 pKa = 3.62LRR251 pKa = 11.84VSTPLAYY258 pKa = 10.41DD259 pKa = 3.18HH260 pKa = 6.88TGVLSVVTRR269 pKa = 11.84YY270 pKa = 9.72PLDD273 pKa = 3.46VTSAGMGVNMQVPLRR288 pKa = 11.84LHH290 pKa = 6.53GVDD293 pKa = 5.51LGLAYY298 pKa = 8.58NTQDD302 pKa = 3.21FSIVDD307 pKa = 4.4GYY309 pKa = 10.16LTLNRR314 pKa = 11.84SQGKK318 pKa = 9.84LEE320 pKa = 4.04EE321 pKa = 4.61LGEE324 pKa = 4.31AVNMNATLVDD334 pKa = 4.55LNDD337 pKa = 4.35GGLQALEE344 pKa = 4.3TDD346 pKa = 4.33LSLEE350 pKa = 4.17KK351 pKa = 10.21PLNAMQCLFEE361 pKa = 6.06HH362 pKa = 6.77EE363 pKa = 4.71GTWRR367 pKa = 11.84WKK369 pKa = 10.18FGVRR373 pKa = 11.84MSTEE377 pKa = 3.69PTITTVRR384 pKa = 11.84VNVHH388 pKa = 5.32STWSVAVQEE397 pKa = 5.02SIAICAITTTTAGAGRR413 pKa = 11.84GQGPRR418 pKa = 11.84VTIPFRR424 pKa = 11.84RR425 pKa = 11.84ADD427 pKa = 3.57LTDD430 pKa = 3.4DD431 pKa = 3.45MMRR434 pKa = 11.84AGFDD438 pKa = 3.25VWKK441 pKa = 10.22SDD443 pKa = 3.91YY444 pKa = 10.42IRR446 pKa = 11.84EE447 pKa = 4.02EE448 pKa = 4.14AFVGTLRR455 pKa = 11.84GLYY458 pKa = 9.82RR459 pKa = 11.84RR460 pKa = 11.84WSDD463 pKa = 2.83KK464 pKa = 10.88HH465 pKa = 7.63LLAEE469 pKa = 5.35FYY471 pKa = 11.23AHH473 pKa = 6.35GRR475 pKa = 11.84LYY477 pKa = 9.38PTEE480 pKa = 4.84DD481 pKa = 4.49DD482 pKa = 3.83IEE484 pKa = 4.52LVLTPRR490 pKa = 11.84STDD493 pKa = 4.95DD494 pKa = 3.44EE495 pKa = 4.89YY496 pKa = 11.65EE497 pKa = 3.96WAGLRR502 pKa = 11.84NVHH505 pKa = 5.2WTVVCCKK512 pKa = 10.53
MM1 pKa = 7.37ATRR4 pKa = 11.84DD5 pKa = 3.35SRR7 pKa = 11.84MTTEE11 pKa = 4.56HH12 pKa = 6.66VIALILAYY20 pKa = 9.68STTLDD25 pKa = 3.19EE26 pKa = 4.9AKK28 pKa = 10.02IHH30 pKa = 6.25EE31 pKa = 5.49EE32 pKa = 3.53ITNLQSAVEE41 pKa = 3.95QLTVRR46 pKa = 11.84LTEE49 pKa = 3.86LTNAVTSLDD58 pKa = 3.4ARR60 pKa = 11.84LTQVEE65 pKa = 4.26QQASKK70 pKa = 11.08LDD72 pKa = 3.85DD73 pKa = 4.67KK74 pKa = 11.4IGILSNRR81 pKa = 11.84IGANTASINFLAGIVDD97 pKa = 4.06ATRR100 pKa = 11.84IDD102 pKa = 4.15AVDD105 pKa = 3.7APLYY109 pKa = 10.36IYY111 pKa = 10.38IVDD114 pKa = 3.74DD115 pKa = 3.55QRR117 pKa = 11.84RR118 pKa = 11.84LAIATGDD125 pKa = 3.62GLFVKK130 pKa = 10.19DD131 pKa = 4.43RR132 pKa = 11.84KK133 pKa = 10.75LNGYY137 pKa = 7.27DD138 pKa = 3.29VRR140 pKa = 11.84SFPPIAVAKK149 pKa = 10.34YY150 pKa = 10.17NDD152 pKa = 3.58ILSFSLSSAPPLDD165 pKa = 3.55IVDD168 pKa = 4.01GKK170 pKa = 10.89LAVSTTSRR178 pKa = 11.84LFITSGKK185 pKa = 10.06LDD187 pKa = 3.55TNSYY191 pKa = 9.64VGSSSVDD198 pKa = 2.95ISGAAAEE205 pKa = 4.42KK206 pKa = 8.81TVSVRR211 pKa = 11.84TINPIIAGQDD221 pKa = 3.54GLSLSLSGVLEE232 pKa = 4.3VGSGLLLSGRR242 pKa = 11.84SWPLTLDD249 pKa = 3.62LRR251 pKa = 11.84VSTPLAYY258 pKa = 10.41DD259 pKa = 3.18HH260 pKa = 6.88TGVLSVVTRR269 pKa = 11.84YY270 pKa = 9.72PLDD273 pKa = 3.46VTSAGMGVNMQVPLRR288 pKa = 11.84LHH290 pKa = 6.53GVDD293 pKa = 5.51LGLAYY298 pKa = 8.58NTQDD302 pKa = 3.21FSIVDD307 pKa = 4.4GYY309 pKa = 10.16LTLNRR314 pKa = 11.84SQGKK318 pKa = 9.84LEE320 pKa = 4.04EE321 pKa = 4.61LGEE324 pKa = 4.31AVNMNATLVDD334 pKa = 4.55LNDD337 pKa = 4.35GGLQALEE344 pKa = 4.3TDD346 pKa = 4.33LSLEE350 pKa = 4.17KK351 pKa = 10.21PLNAMQCLFEE361 pKa = 6.06HH362 pKa = 6.77EE363 pKa = 4.71GTWRR367 pKa = 11.84WKK369 pKa = 10.18FGVRR373 pKa = 11.84MSTEE377 pKa = 3.69PTITTVRR384 pKa = 11.84VNVHH388 pKa = 5.32STWSVAVQEE397 pKa = 5.02SIAICAITTTTAGAGRR413 pKa = 11.84GQGPRR418 pKa = 11.84VTIPFRR424 pKa = 11.84RR425 pKa = 11.84ADD427 pKa = 3.57LTDD430 pKa = 3.4DD431 pKa = 3.45MMRR434 pKa = 11.84AGFDD438 pKa = 3.25VWKK441 pKa = 10.22SDD443 pKa = 3.91YY444 pKa = 10.42IRR446 pKa = 11.84EE447 pKa = 4.02EE448 pKa = 4.14AFVGTLRR455 pKa = 11.84GLYY458 pKa = 9.82RR459 pKa = 11.84RR460 pKa = 11.84WSDD463 pKa = 2.83KK464 pKa = 10.88HH465 pKa = 7.63LLAEE469 pKa = 5.35FYY471 pKa = 11.23AHH473 pKa = 6.35GRR475 pKa = 11.84LYY477 pKa = 9.38PTEE480 pKa = 4.84DD481 pKa = 4.49DD482 pKa = 3.83IEE484 pKa = 4.52LVLTPRR490 pKa = 11.84STDD493 pKa = 4.95DD494 pKa = 3.44EE495 pKa = 4.89YY496 pKa = 11.65EE497 pKa = 3.96WAGLRR502 pKa = 11.84NVHH505 pKa = 5.2WTVVCCKK512 pKa = 10.53
Molecular weight: 56.04 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|E7DDL0|E7DDL0_9REOV Reovirus virion structural protein OS=Grass carp reovirus OX=128987 GN=VP6 PE=4 SV=1
MM1 pKa = 7.34YY2 pKa = 10.44LEE4 pKa = 4.99LFIAVMWHH12 pKa = 6.33FMLAFCAYY20 pKa = 7.91VWYY23 pKa = 10.07IHH25 pKa = 6.71GPTLRR30 pKa = 11.84RR31 pKa = 11.84LYY33 pKa = 10.58QRR35 pKa = 11.84LTATTANAICQTDD48 pKa = 3.38STYY51 pKa = 11.28VCTATVSTMTDD62 pKa = 3.21DD63 pKa = 3.63LTNASSSASTSSTASSATTSFVALPRR89 pKa = 11.84LSLDD93 pKa = 3.76DD94 pKa = 5.08LKK96 pKa = 10.3THH98 pKa = 6.96SKK100 pKa = 10.14FRR102 pKa = 11.84WLWFSNNPLTIDD114 pKa = 4.19LSTDD118 pKa = 3.05EE119 pKa = 4.83LTTPFNSPQLHH130 pKa = 6.76DD131 pKa = 3.94VLFMPQYY138 pKa = 9.93TLAAYY143 pKa = 8.75PYY145 pKa = 7.7MHH147 pKa = 7.43RR148 pKa = 11.84SLHH151 pKa = 5.41LALYY155 pKa = 10.05FRR157 pKa = 11.84FGYY160 pKa = 9.58NMRR163 pKa = 11.84YY164 pKa = 9.64LLRR167 pKa = 11.84EE168 pKa = 3.98HH169 pKa = 7.1PGIPPLRR176 pKa = 11.84VTCPSMRR183 pKa = 11.84VLFMDD188 pKa = 3.72KK189 pKa = 10.22QRR191 pKa = 11.84NKK193 pKa = 10.7LEE195 pKa = 4.17EE196 pKa = 4.39PVCSICRR203 pKa = 11.84LWIVDD208 pKa = 4.19GPQLPIGNCGRR219 pKa = 11.84KK220 pKa = 9.6YY221 pKa = 10.11KK222 pKa = 10.33VHH224 pKa = 6.96EE225 pKa = 4.15IFNAFSLEE233 pKa = 3.72PHH235 pKa = 6.81HH236 pKa = 6.82IVNAFDD242 pKa = 4.0VIPIISLLKK251 pKa = 10.42CLQLDD256 pKa = 3.57ISMLAGLSLTALTQLSEE273 pKa = 4.18LSRR276 pKa = 11.84DD277 pKa = 3.61PDD279 pKa = 3.47VTPAGFLAAIQFDD292 pKa = 4.28GHH294 pKa = 6.4TSTVDD299 pKa = 3.15CRR301 pKa = 11.84GDD303 pKa = 3.1RR304 pKa = 11.84LYY306 pKa = 11.09SHH308 pKa = 7.48PMLLILFALFCYY320 pKa = 9.99RR321 pKa = 11.84RR322 pKa = 11.84RR323 pKa = 11.84SRR325 pKa = 11.84VYY327 pKa = 9.98FRR329 pKa = 11.84EE330 pKa = 4.27CYY332 pKa = 10.06LRR334 pKa = 11.84DD335 pKa = 3.55YY336 pKa = 11.19RR337 pKa = 11.84RR338 pKa = 11.84LTSSLPDD345 pKa = 3.21MVPYY349 pKa = 10.74SRR351 pKa = 11.84TPGKK355 pKa = 10.49AKK357 pKa = 10.51AKK359 pKa = 10.56SPP361 pKa = 3.64
MM1 pKa = 7.34YY2 pKa = 10.44LEE4 pKa = 4.99LFIAVMWHH12 pKa = 6.33FMLAFCAYY20 pKa = 7.91VWYY23 pKa = 10.07IHH25 pKa = 6.71GPTLRR30 pKa = 11.84RR31 pKa = 11.84LYY33 pKa = 10.58QRR35 pKa = 11.84LTATTANAICQTDD48 pKa = 3.38STYY51 pKa = 11.28VCTATVSTMTDD62 pKa = 3.21DD63 pKa = 3.63LTNASSSASTSSTASSATTSFVALPRR89 pKa = 11.84LSLDD93 pKa = 3.76DD94 pKa = 5.08LKK96 pKa = 10.3THH98 pKa = 6.96SKK100 pKa = 10.14FRR102 pKa = 11.84WLWFSNNPLTIDD114 pKa = 4.19LSTDD118 pKa = 3.05EE119 pKa = 4.83LTTPFNSPQLHH130 pKa = 6.76DD131 pKa = 3.94VLFMPQYY138 pKa = 9.93TLAAYY143 pKa = 8.75PYY145 pKa = 7.7MHH147 pKa = 7.43RR148 pKa = 11.84SLHH151 pKa = 5.41LALYY155 pKa = 10.05FRR157 pKa = 11.84FGYY160 pKa = 9.58NMRR163 pKa = 11.84YY164 pKa = 9.64LLRR167 pKa = 11.84EE168 pKa = 3.98HH169 pKa = 7.1PGIPPLRR176 pKa = 11.84VTCPSMRR183 pKa = 11.84VLFMDD188 pKa = 3.72KK189 pKa = 10.22QRR191 pKa = 11.84NKK193 pKa = 10.7LEE195 pKa = 4.17EE196 pKa = 4.39PVCSICRR203 pKa = 11.84LWIVDD208 pKa = 4.19GPQLPIGNCGRR219 pKa = 11.84KK220 pKa = 9.6YY221 pKa = 10.11KK222 pKa = 10.33VHH224 pKa = 6.96EE225 pKa = 4.15IFNAFSLEE233 pKa = 3.72PHH235 pKa = 6.81HH236 pKa = 6.82IVNAFDD242 pKa = 4.0VIPIISLLKK251 pKa = 10.42CLQLDD256 pKa = 3.57ISMLAGLSLTALTQLSEE273 pKa = 4.18LSRR276 pKa = 11.84DD277 pKa = 3.61PDD279 pKa = 3.47VTPAGFLAAIQFDD292 pKa = 4.28GHH294 pKa = 6.4TSTVDD299 pKa = 3.15CRR301 pKa = 11.84GDD303 pKa = 3.1RR304 pKa = 11.84LYY306 pKa = 11.09SHH308 pKa = 7.48PMLLILFALFCYY320 pKa = 9.99RR321 pKa = 11.84RR322 pKa = 11.84RR323 pKa = 11.84SRR325 pKa = 11.84VYY327 pKa = 9.98FRR329 pKa = 11.84EE330 pKa = 4.27CYY332 pKa = 10.06LRR334 pKa = 11.84DD335 pKa = 3.55YY336 pKa = 11.19RR337 pKa = 11.84RR338 pKa = 11.84LTSSLPDD345 pKa = 3.21MVPYY349 pKa = 10.74SRR351 pKa = 11.84TPGKK355 pKa = 10.49AKK357 pKa = 10.51AKK359 pKa = 10.56SPP361 pKa = 3.64
Molecular weight: 41.31 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
7837 |
310 |
1294 |
712.5 |
79.02 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.332 ± 0.434 | 1.544 ± 0.148 |
6.278 ± 0.234 | 3.879 ± 0.273 |
3.535 ± 0.238 | 6.087 ± 0.489 |
2.131 ± 0.262 | 5.627 ± 0.278 |
3.241 ± 0.172 | 10.08 ± 0.433 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.871 ± 0.155 | 3.828 ± 0.255 |
5.678 ± 0.253 | 3.33 ± 0.281 |
6.24 ± 0.39 | 7.962 ± 0.468 |
7.324 ± 0.586 | 6.929 ± 0.441 |
1.799 ± 0.19 | 3.305 ± 0.232 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
