
Planctomycetes bacterium Pan265
Taxonomy: cellular organisms; Bacteria; PVC group; Planctomycetes; unclassified Planctomycetes
Average proteome isoelectric point is 5.79
Get precalculated fractions of proteins
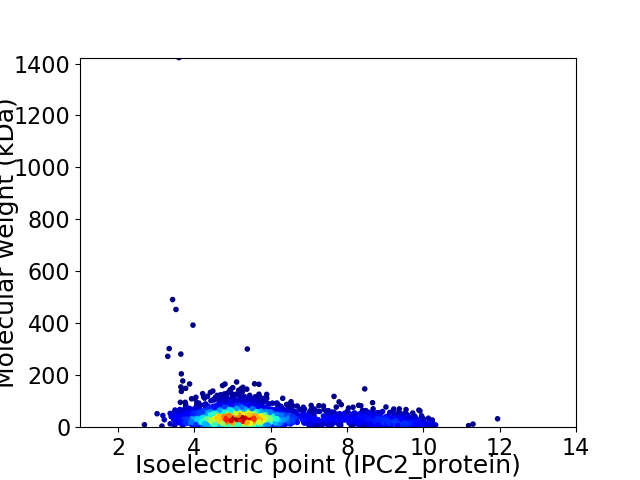
Virtual 2D-PAGE plot for 2855 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A518C040|A0A518C040_9BACT Uncharacterized protein OS=Planctomycetes bacterium Pan265 OX=2527982 GN=Pan265_24570 PE=4 SV=1
MM1 pKa = 7.18TRR3 pKa = 11.84ITSGGVLASTLILALTTTTHH23 pKa = 6.33ADD25 pKa = 2.93IYY27 pKa = 10.68QWTASRR33 pKa = 11.84GIYY36 pKa = 10.05QSTTLTPDD44 pKa = 3.32GAGKK48 pKa = 8.04TAEE51 pKa = 4.88PYY53 pKa = 11.15ADD55 pKa = 3.51LQGLNLYY62 pKa = 9.7KK63 pKa = 10.55AYY65 pKa = 10.58LRR67 pKa = 11.84EE68 pKa = 4.25ADD70 pKa = 3.65LTQANLSNANLADD83 pKa = 4.15AYY85 pKa = 10.81LYY87 pKa = 11.01NATLTNTDD95 pKa = 3.92LSDD98 pKa = 5.43AIITEE103 pKa = 4.41ANFGSTTDD111 pKa = 3.92FGFTATQLYY120 pKa = 8.16STASYY125 pKa = 10.7KK126 pKa = 10.81NKK128 pKa = 10.0NLAGIKK134 pKa = 10.05LYY136 pKa = 11.34SNDD139 pKa = 3.19LTGWNLAGQNLASASFYY156 pKa = 11.23NATLTNTDD164 pKa = 3.92LSDD167 pKa = 5.43AIITEE172 pKa = 4.41ANFGSTTDD180 pKa = 3.27SGFTAAQLYY189 pKa = 7.97STASYY194 pKa = 10.7KK195 pKa = 10.81NKK197 pKa = 10.07NLAGIDD203 pKa = 3.65LRR205 pKa = 11.84YY206 pKa = 10.39NDD208 pKa = 3.64LTGWNLAGQNLTDD221 pKa = 5.37ADD223 pKa = 4.06FDD225 pKa = 4.76DD226 pKa = 5.98ANLTNTDD233 pKa = 4.45LNNADD238 pKa = 4.02LTDD241 pKa = 4.16ANLDD245 pKa = 3.77YY246 pKa = 11.77ANLTNTDD253 pKa = 3.72LSDD256 pKa = 5.43AIITEE261 pKa = 4.41ANFGSTTDD269 pKa = 3.18SGFTAGQLYY278 pKa = 8.26STASYY283 pKa = 10.7KK284 pKa = 10.81NKK286 pKa = 10.07NLAGIDD292 pKa = 3.65LRR294 pKa = 11.84YY295 pKa = 10.39NDD297 pKa = 3.57LTAWNLAGQNLTDD310 pKa = 4.59AYY312 pKa = 10.83FGLANLANTDD322 pKa = 3.93LSDD325 pKa = 4.43ALITGADD332 pKa = 4.58FYY334 pKa = 10.87RR335 pKa = 11.84TTDD338 pKa = 2.95SGFTAAQLYY347 pKa = 7.97STASYY352 pKa = 10.71KK353 pKa = 10.81NKK355 pKa = 10.13NLTGIDD361 pKa = 3.5LGYY364 pKa = 11.04NDD366 pKa = 3.71LTGWNLAGQNLTNTDD381 pKa = 3.87LNNANLTDD389 pKa = 3.91AYY391 pKa = 10.62LYY393 pKa = 10.98NATLTNTDD401 pKa = 3.92LSDD404 pKa = 5.43AIITEE409 pKa = 4.41ANFGSTTDD417 pKa = 3.24SGFTATQLYY426 pKa = 8.16STASYY431 pKa = 10.71KK432 pKa = 10.81NKK434 pKa = 10.06NLTGIKK440 pKa = 10.03LYY442 pKa = 11.31SNDD445 pKa = 3.19LTGWNLAGQNLTDD458 pKa = 3.75TDD460 pKa = 4.05LDD462 pKa = 4.14YY463 pKa = 11.89ANLTNTDD470 pKa = 4.0LNNANLTDD478 pKa = 4.27ANLDD482 pKa = 3.74YY483 pKa = 11.77ANLTNTDD490 pKa = 3.94LSDD493 pKa = 3.88ALITGADD500 pKa = 4.58FYY502 pKa = 10.87RR503 pKa = 11.84TTDD506 pKa = 2.91SGFTAGQLYY515 pKa = 8.26STASYY520 pKa = 10.7KK521 pKa = 10.81NKK523 pKa = 10.07NLAGIDD529 pKa = 3.56LGYY532 pKa = 11.05NDD534 pKa = 4.48LTDD537 pKa = 3.5WDD539 pKa = 4.6LSNLNLQSVVFNHH552 pKa = 6.94ANLHH556 pKa = 5.22NTDD559 pKa = 4.07FTASDD564 pKa = 3.89LRR566 pKa = 11.84RR567 pKa = 11.84SHH569 pKa = 6.58YY570 pKa = 10.97YY571 pKa = 9.49IDD573 pKa = 3.51RR574 pKa = 11.84TSGNRR579 pKa = 11.84MSQRR583 pKa = 11.84LAGVRR588 pKa = 11.84NAIHH592 pKa = 6.98PDD594 pKa = 3.09GRR596 pKa = 11.84VKK598 pKa = 10.93GLILQSSEE606 pKa = 4.03TLRR609 pKa = 11.84IWDD612 pKa = 4.86DD613 pKa = 3.61NPLDD617 pKa = 3.93EE618 pKa = 5.5SEE620 pKa = 4.1PAIPITIEE628 pKa = 3.66QEE630 pKa = 4.1LTIDD634 pKa = 3.57PAATLRR640 pKa = 11.84LVFEE644 pKa = 4.99DD645 pKa = 4.11NQWGSIIQFADD656 pKa = 3.29PDD658 pKa = 4.3TPVALAGTLLLEE670 pKa = 4.75FDD672 pKa = 5.5DD673 pKa = 5.96GLDD676 pKa = 3.71LQSLQITTYY685 pKa = 11.24QLFDD689 pKa = 3.46WTNADD694 pKa = 2.71ITGAFDD700 pKa = 3.94TILYY704 pKa = 8.72NGPGTFDD711 pKa = 3.36TSSLMATGEE720 pKa = 4.07VTYY723 pKa = 9.81FSATLLPADD732 pKa = 4.43ANLDD736 pKa = 3.52GSVDD740 pKa = 3.8MNDD743 pKa = 4.36LSILAANFGLPGVFGFTQGDD763 pKa = 3.93FNGDD767 pKa = 3.33NTVDD771 pKa = 4.61LLDD774 pKa = 4.22LSILASHH781 pKa = 7.57FGNTAIPEE789 pKa = 4.17PAAVTILLTLLPLSRR804 pKa = 11.84RR805 pKa = 11.84RR806 pKa = 11.84SAA808 pKa = 3.98
MM1 pKa = 7.18TRR3 pKa = 11.84ITSGGVLASTLILALTTTTHH23 pKa = 6.33ADD25 pKa = 2.93IYY27 pKa = 10.68QWTASRR33 pKa = 11.84GIYY36 pKa = 10.05QSTTLTPDD44 pKa = 3.32GAGKK48 pKa = 8.04TAEE51 pKa = 4.88PYY53 pKa = 11.15ADD55 pKa = 3.51LQGLNLYY62 pKa = 9.7KK63 pKa = 10.55AYY65 pKa = 10.58LRR67 pKa = 11.84EE68 pKa = 4.25ADD70 pKa = 3.65LTQANLSNANLADD83 pKa = 4.15AYY85 pKa = 10.81LYY87 pKa = 11.01NATLTNTDD95 pKa = 3.92LSDD98 pKa = 5.43AIITEE103 pKa = 4.41ANFGSTTDD111 pKa = 3.92FGFTATQLYY120 pKa = 8.16STASYY125 pKa = 10.7KK126 pKa = 10.81NKK128 pKa = 10.0NLAGIKK134 pKa = 10.05LYY136 pKa = 11.34SNDD139 pKa = 3.19LTGWNLAGQNLASASFYY156 pKa = 11.23NATLTNTDD164 pKa = 3.92LSDD167 pKa = 5.43AIITEE172 pKa = 4.41ANFGSTTDD180 pKa = 3.27SGFTAAQLYY189 pKa = 7.97STASYY194 pKa = 10.7KK195 pKa = 10.81NKK197 pKa = 10.07NLAGIDD203 pKa = 3.65LRR205 pKa = 11.84YY206 pKa = 10.39NDD208 pKa = 3.64LTGWNLAGQNLTDD221 pKa = 5.37ADD223 pKa = 4.06FDD225 pKa = 4.76DD226 pKa = 5.98ANLTNTDD233 pKa = 4.45LNNADD238 pKa = 4.02LTDD241 pKa = 4.16ANLDD245 pKa = 3.77YY246 pKa = 11.77ANLTNTDD253 pKa = 3.72LSDD256 pKa = 5.43AIITEE261 pKa = 4.41ANFGSTTDD269 pKa = 3.18SGFTAGQLYY278 pKa = 8.26STASYY283 pKa = 10.7KK284 pKa = 10.81NKK286 pKa = 10.07NLAGIDD292 pKa = 3.65LRR294 pKa = 11.84YY295 pKa = 10.39NDD297 pKa = 3.57LTAWNLAGQNLTDD310 pKa = 4.59AYY312 pKa = 10.83FGLANLANTDD322 pKa = 3.93LSDD325 pKa = 4.43ALITGADD332 pKa = 4.58FYY334 pKa = 10.87RR335 pKa = 11.84TTDD338 pKa = 2.95SGFTAAQLYY347 pKa = 7.97STASYY352 pKa = 10.71KK353 pKa = 10.81NKK355 pKa = 10.13NLTGIDD361 pKa = 3.5LGYY364 pKa = 11.04NDD366 pKa = 3.71LTGWNLAGQNLTNTDD381 pKa = 3.87LNNANLTDD389 pKa = 3.91AYY391 pKa = 10.62LYY393 pKa = 10.98NATLTNTDD401 pKa = 3.92LSDD404 pKa = 5.43AIITEE409 pKa = 4.41ANFGSTTDD417 pKa = 3.24SGFTATQLYY426 pKa = 8.16STASYY431 pKa = 10.71KK432 pKa = 10.81NKK434 pKa = 10.06NLTGIKK440 pKa = 10.03LYY442 pKa = 11.31SNDD445 pKa = 3.19LTGWNLAGQNLTDD458 pKa = 3.75TDD460 pKa = 4.05LDD462 pKa = 4.14YY463 pKa = 11.89ANLTNTDD470 pKa = 4.0LNNANLTDD478 pKa = 4.27ANLDD482 pKa = 3.74YY483 pKa = 11.77ANLTNTDD490 pKa = 3.94LSDD493 pKa = 3.88ALITGADD500 pKa = 4.58FYY502 pKa = 10.87RR503 pKa = 11.84TTDD506 pKa = 2.91SGFTAGQLYY515 pKa = 8.26STASYY520 pKa = 10.7KK521 pKa = 10.81NKK523 pKa = 10.07NLAGIDD529 pKa = 3.56LGYY532 pKa = 11.05NDD534 pKa = 4.48LTDD537 pKa = 3.5WDD539 pKa = 4.6LSNLNLQSVVFNHH552 pKa = 6.94ANLHH556 pKa = 5.22NTDD559 pKa = 4.07FTASDD564 pKa = 3.89LRR566 pKa = 11.84RR567 pKa = 11.84SHH569 pKa = 6.58YY570 pKa = 10.97YY571 pKa = 9.49IDD573 pKa = 3.51RR574 pKa = 11.84TSGNRR579 pKa = 11.84MSQRR583 pKa = 11.84LAGVRR588 pKa = 11.84NAIHH592 pKa = 6.98PDD594 pKa = 3.09GRR596 pKa = 11.84VKK598 pKa = 10.93GLILQSSEE606 pKa = 4.03TLRR609 pKa = 11.84IWDD612 pKa = 4.86DD613 pKa = 3.61NPLDD617 pKa = 3.93EE618 pKa = 5.5SEE620 pKa = 4.1PAIPITIEE628 pKa = 3.66QEE630 pKa = 4.1LTIDD634 pKa = 3.57PAATLRR640 pKa = 11.84LVFEE644 pKa = 4.99DD645 pKa = 4.11NQWGSIIQFADD656 pKa = 3.29PDD658 pKa = 4.3TPVALAGTLLLEE670 pKa = 4.75FDD672 pKa = 5.5DD673 pKa = 5.96GLDD676 pKa = 3.71LQSLQITTYY685 pKa = 11.24QLFDD689 pKa = 3.46WTNADD694 pKa = 2.71ITGAFDD700 pKa = 3.94TILYY704 pKa = 8.72NGPGTFDD711 pKa = 3.36TSSLMATGEE720 pKa = 4.07VTYY723 pKa = 9.81FSATLLPADD732 pKa = 4.43ANLDD736 pKa = 3.52GSVDD740 pKa = 3.8MNDD743 pKa = 4.36LSILAANFGLPGVFGFTQGDD763 pKa = 3.93FNGDD767 pKa = 3.33NTVDD771 pKa = 4.61LLDD774 pKa = 4.22LSILASHH781 pKa = 7.57FGNTAIPEE789 pKa = 4.17PAAVTILLTLLPLSRR804 pKa = 11.84RR805 pKa = 11.84RR806 pKa = 11.84SAA808 pKa = 3.98
Molecular weight: 87.19 kDa
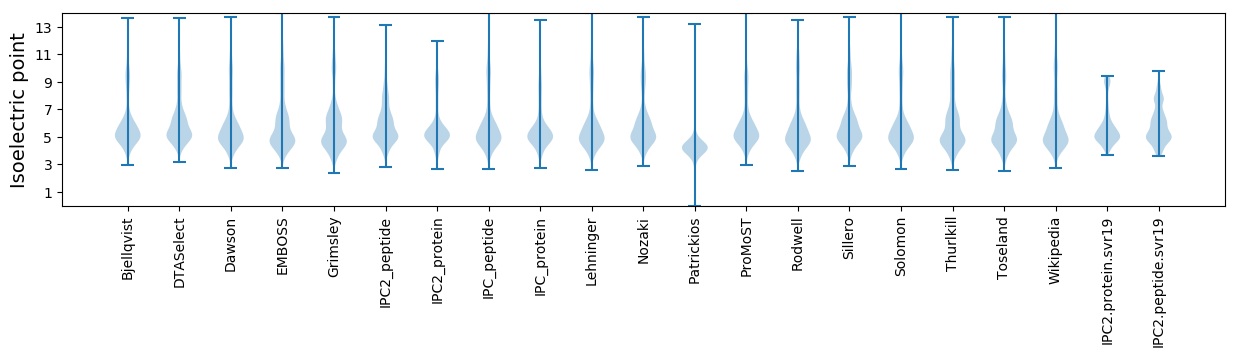
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A518BXZ9|A0A518BXZ9_9BACT Uncharacterized protein OS=Planctomycetes bacterium Pan265 OX=2527982 GN=Pan265_16960 PE=4 SV=1
MM1 pKa = 6.78STHH4 pKa = 4.96YY5 pKa = 9.85PKK7 pKa = 10.32RR8 pKa = 11.84RR9 pKa = 11.84SRR11 pKa = 11.84IKK13 pKa = 10.22RR14 pKa = 11.84SRR16 pKa = 11.84MWGFRR21 pKa = 11.84ARR23 pKa = 11.84MKK25 pKa = 9.78TKK27 pKa = 10.24QGRR30 pKa = 11.84KK31 pKa = 7.48MINRR35 pKa = 11.84KK36 pKa = 9.07RR37 pKa = 11.84RR38 pKa = 11.84VGRR41 pKa = 11.84SVNVRR46 pKa = 11.84HH47 pKa = 6.13NFF49 pKa = 3.25
MM1 pKa = 6.78STHH4 pKa = 4.96YY5 pKa = 9.85PKK7 pKa = 10.32RR8 pKa = 11.84RR9 pKa = 11.84SRR11 pKa = 11.84IKK13 pKa = 10.22RR14 pKa = 11.84SRR16 pKa = 11.84MWGFRR21 pKa = 11.84ARR23 pKa = 11.84MKK25 pKa = 9.78TKK27 pKa = 10.24QGRR30 pKa = 11.84KK31 pKa = 7.48MINRR35 pKa = 11.84KK36 pKa = 9.07RR37 pKa = 11.84RR38 pKa = 11.84VGRR41 pKa = 11.84SVNVRR46 pKa = 11.84HH47 pKa = 6.13NFF49 pKa = 3.25
Molecular weight: 6.14 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1052711 |
32 |
13493 |
368.7 |
40.34 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.529 ± 0.048 | 0.876 ± 0.021 |
6.715 ± 0.074 | 6.08 ± 0.057 |
3.327 ± 0.032 | 8.363 ± 0.062 |
2.294 ± 0.035 | 5.042 ± 0.04 |
2.556 ± 0.048 | 10.215 ± 0.051 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.263 ± 0.036 | 3.047 ± 0.05 |
5.189 ± 0.042 | 3.497 ± 0.032 |
6.961 ± 0.08 | 5.516 ± 0.041 |
6.067 ± 0.08 | 7.497 ± 0.044 |
1.5 ± 0.026 | 2.466 ± 0.028 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
