
Paenibacillus curdlanolyticus YK9
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Bacillales; Paenibacillaceae; Paenibacillus; Paenibacillus curdlanolyticus
Average proteome isoelectric point is 6.31
Get precalculated fractions of proteins
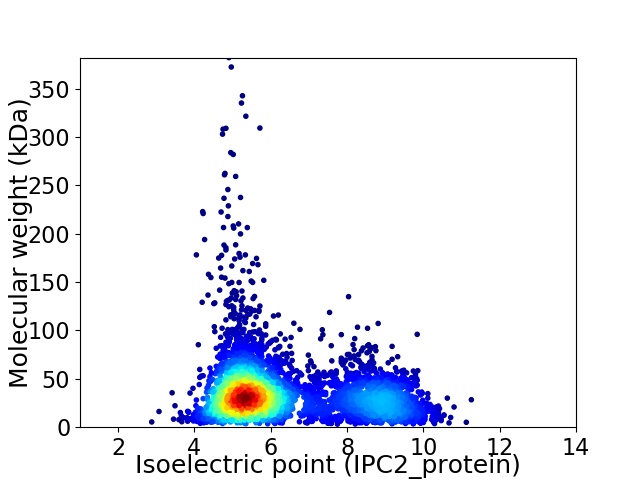
Virtual 2D-PAGE plot for 4803 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|E0IGP1|E0IGP1_9BACL Mutator family transposase (Fragment) OS=Paenibacillus curdlanolyticus YK9 OX=717606 GN=PaecuDRAFT_4836 PE=3 SV=1
MM1 pKa = 8.33VIMIVGCGSQDD12 pKa = 3.45PADD15 pKa = 3.89NDD17 pKa = 3.18LRR19 pKa = 11.84AAEE22 pKa = 3.96QPIVEE27 pKa = 4.36EE28 pKa = 4.46TSASSEE34 pKa = 4.19VVANEE39 pKa = 3.63SAEE42 pKa = 4.11PKK44 pKa = 8.78EE45 pKa = 4.03QEE47 pKa = 3.92IAEE50 pKa = 4.25TDD52 pKa = 3.48AVTVNYY58 pKa = 9.24AAQYY62 pKa = 8.05NGQWVTLIKK71 pKa = 10.32PEE73 pKa = 4.27RR74 pKa = 11.84CGTCKK79 pKa = 9.45NTLNIEE85 pKa = 3.83IDD87 pKa = 3.74SANQATVSIQQASDD101 pKa = 3.21HH102 pKa = 5.84GTHH105 pKa = 7.07LAGIDD110 pKa = 3.57FTVQLDD116 pKa = 4.27GQGSGAFTFDD126 pKa = 3.14EE127 pKa = 5.39DD128 pKa = 3.81GWFHH132 pKa = 5.99QGKK135 pKa = 8.24GTISLQSDD143 pKa = 4.13VIEE146 pKa = 4.21VTITEE151 pKa = 4.4MKK153 pKa = 10.53DD154 pKa = 3.18NPDD157 pKa = 3.33ADD159 pKa = 3.99EE160 pKa = 4.28VSNTYY165 pKa = 10.84NIFSGTQQFYY175 pKa = 10.11RR176 pKa = 11.84PEE178 pKa = 4.0VNRR181 pKa = 11.84RR182 pKa = 11.84AIMAKK187 pKa = 10.32LMEE190 pKa = 4.97DD191 pKa = 3.32TDD193 pKa = 3.63IDD195 pKa = 3.76ALEE198 pKa = 4.28FKK200 pKa = 11.28YY201 pKa = 10.66EE202 pKa = 3.97DD203 pKa = 3.56EE204 pKa = 4.12KK205 pKa = 11.61GYY207 pKa = 9.81WYY209 pKa = 10.5YY210 pKa = 11.11CAVIDD215 pKa = 4.52PNSEE219 pKa = 4.06GEE221 pKa = 4.09PLCYY225 pKa = 10.48GIDD228 pKa = 3.87PYY230 pKa = 11.02SGKK233 pKa = 10.38YY234 pKa = 10.03FDD236 pKa = 6.15DD237 pKa = 3.42ISGGYY242 pKa = 8.67EE243 pKa = 3.63GNIYY247 pKa = 10.17SDD249 pKa = 3.41HH250 pKa = 6.63SVPDD254 pKa = 3.89FDD256 pKa = 4.67EE257 pKa = 4.05VAAFVKK263 pKa = 10.56QKK265 pKa = 11.13LNLEE269 pKa = 4.3GDD271 pKa = 3.92TQFNGSDD278 pKa = 3.2YY279 pKa = 11.19GLYY282 pKa = 9.51IVQVGEE288 pKa = 4.01QEE290 pKa = 4.16FEE292 pKa = 4.13YY293 pKa = 11.03DD294 pKa = 3.93SDD296 pKa = 3.77TGDD299 pKa = 3.9LFDD302 pKa = 5.49SKK304 pKa = 7.67THH306 pKa = 6.46KK307 pKa = 10.89LLDD310 pKa = 4.71GIGLIEE316 pKa = 4.06PTQQ319 pKa = 3.3
MM1 pKa = 8.33VIMIVGCGSQDD12 pKa = 3.45PADD15 pKa = 3.89NDD17 pKa = 3.18LRR19 pKa = 11.84AAEE22 pKa = 3.96QPIVEE27 pKa = 4.36EE28 pKa = 4.46TSASSEE34 pKa = 4.19VVANEE39 pKa = 3.63SAEE42 pKa = 4.11PKK44 pKa = 8.78EE45 pKa = 4.03QEE47 pKa = 3.92IAEE50 pKa = 4.25TDD52 pKa = 3.48AVTVNYY58 pKa = 9.24AAQYY62 pKa = 8.05NGQWVTLIKK71 pKa = 10.32PEE73 pKa = 4.27RR74 pKa = 11.84CGTCKK79 pKa = 9.45NTLNIEE85 pKa = 3.83IDD87 pKa = 3.74SANQATVSIQQASDD101 pKa = 3.21HH102 pKa = 5.84GTHH105 pKa = 7.07LAGIDD110 pKa = 3.57FTVQLDD116 pKa = 4.27GQGSGAFTFDD126 pKa = 3.14EE127 pKa = 5.39DD128 pKa = 3.81GWFHH132 pKa = 5.99QGKK135 pKa = 8.24GTISLQSDD143 pKa = 4.13VIEE146 pKa = 4.21VTITEE151 pKa = 4.4MKK153 pKa = 10.53DD154 pKa = 3.18NPDD157 pKa = 3.33ADD159 pKa = 3.99EE160 pKa = 4.28VSNTYY165 pKa = 10.84NIFSGTQQFYY175 pKa = 10.11RR176 pKa = 11.84PEE178 pKa = 4.0VNRR181 pKa = 11.84RR182 pKa = 11.84AIMAKK187 pKa = 10.32LMEE190 pKa = 4.97DD191 pKa = 3.32TDD193 pKa = 3.63IDD195 pKa = 3.76ALEE198 pKa = 4.28FKK200 pKa = 11.28YY201 pKa = 10.66EE202 pKa = 3.97DD203 pKa = 3.56EE204 pKa = 4.12KK205 pKa = 11.61GYY207 pKa = 9.81WYY209 pKa = 10.5YY210 pKa = 11.11CAVIDD215 pKa = 4.52PNSEE219 pKa = 4.06GEE221 pKa = 4.09PLCYY225 pKa = 10.48GIDD228 pKa = 3.87PYY230 pKa = 11.02SGKK233 pKa = 10.38YY234 pKa = 10.03FDD236 pKa = 6.15DD237 pKa = 3.42ISGGYY242 pKa = 8.67EE243 pKa = 3.63GNIYY247 pKa = 10.17SDD249 pKa = 3.41HH250 pKa = 6.63SVPDD254 pKa = 3.89FDD256 pKa = 4.67EE257 pKa = 4.05VAAFVKK263 pKa = 10.56QKK265 pKa = 11.13LNLEE269 pKa = 4.3GDD271 pKa = 3.92TQFNGSDD278 pKa = 3.2YY279 pKa = 11.19GLYY282 pKa = 9.51IVQVGEE288 pKa = 4.01QEE290 pKa = 4.16FEE292 pKa = 4.13YY293 pKa = 11.03DD294 pKa = 3.93SDD296 pKa = 3.77TGDD299 pKa = 3.9LFDD302 pKa = 5.49SKK304 pKa = 7.67THH306 pKa = 6.46KK307 pKa = 10.89LLDD310 pKa = 4.71GIGLIEE316 pKa = 4.06PTQQ319 pKa = 3.3
Molecular weight: 35.28 kDa
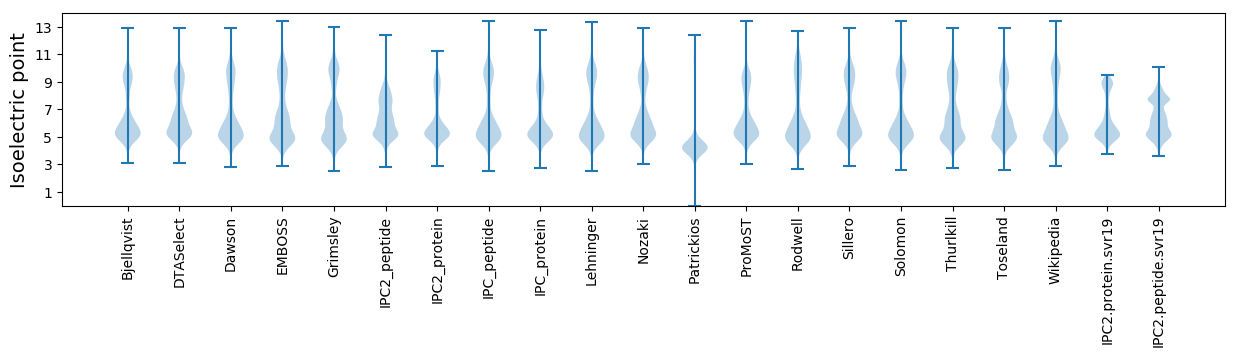
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|E0IBI4|E0IBI4_9BACL MATE efflux family protein OS=Paenibacillus curdlanolyticus YK9 OX=717606 GN=PaecuDRAFT_3023 PE=4 SV=1
MM1 pKa = 8.05PKK3 pKa = 8.13TQKK6 pKa = 10.95ASFPVAQNAMTPLYY20 pKa = 10.45FRR22 pKa = 11.84ACVDD26 pKa = 4.26SIARR30 pKa = 11.84TAMSSLLHH38 pKa = 6.49AATPILARR46 pKa = 11.84HH47 pKa = 5.2TVLARR52 pKa = 11.84HH53 pKa = 6.13AVPSRR58 pKa = 11.84HH59 pKa = 5.94AVLARR64 pKa = 11.84HH65 pKa = 6.01AVLARR70 pKa = 11.84HH71 pKa = 5.97AVLARR76 pKa = 11.84HH77 pKa = 5.42TVLARR82 pKa = 11.84HH83 pKa = 6.15AVPARR88 pKa = 11.84HH89 pKa = 6.08AVLARR94 pKa = 11.84HH95 pKa = 6.09AVPSRR100 pKa = 11.84HH101 pKa = 5.94AVLARR106 pKa = 11.84HH107 pKa = 6.01AVLARR112 pKa = 11.84HH113 pKa = 6.01AVLARR118 pKa = 11.84HH119 pKa = 6.09AVPSRR124 pKa = 11.84HH125 pKa = 5.94AVLARR130 pKa = 11.84HH131 pKa = 6.15AVPARR136 pKa = 11.84HH137 pKa = 5.9AVPSRR142 pKa = 11.84RR143 pKa = 11.84AVLARR148 pKa = 11.84HH149 pKa = 5.87AVLARR154 pKa = 11.84HH155 pKa = 6.01AVLARR160 pKa = 11.84HH161 pKa = 6.01AVLARR166 pKa = 11.84HH167 pKa = 5.97AVLARR172 pKa = 11.84HH173 pKa = 5.56TVPARR178 pKa = 11.84HH179 pKa = 5.96AVLARR184 pKa = 11.84HH185 pKa = 6.01AVLARR190 pKa = 11.84HH191 pKa = 6.09AVPSRR196 pKa = 11.84HH197 pKa = 5.63AVPSRR202 pKa = 11.84RR203 pKa = 11.84AVLARR208 pKa = 11.84HH209 pKa = 5.87AVLARR214 pKa = 11.84HH215 pKa = 5.63TVPSRR220 pKa = 11.84HH221 pKa = 5.97AVLSLTSAIKK231 pKa = 10.23PSTARR236 pKa = 11.84AHH238 pKa = 6.41RR239 pKa = 11.84RR240 pKa = 11.84LPSAIASPTASAQNRR255 pKa = 11.84VSLSRR260 pKa = 11.84RR261 pKa = 11.84GFF263 pKa = 3.39
MM1 pKa = 8.05PKK3 pKa = 8.13TQKK6 pKa = 10.95ASFPVAQNAMTPLYY20 pKa = 10.45FRR22 pKa = 11.84ACVDD26 pKa = 4.26SIARR30 pKa = 11.84TAMSSLLHH38 pKa = 6.49AATPILARR46 pKa = 11.84HH47 pKa = 5.2TVLARR52 pKa = 11.84HH53 pKa = 6.13AVPSRR58 pKa = 11.84HH59 pKa = 5.94AVLARR64 pKa = 11.84HH65 pKa = 6.01AVLARR70 pKa = 11.84HH71 pKa = 5.97AVLARR76 pKa = 11.84HH77 pKa = 5.42TVLARR82 pKa = 11.84HH83 pKa = 6.15AVPARR88 pKa = 11.84HH89 pKa = 6.08AVLARR94 pKa = 11.84HH95 pKa = 6.09AVPSRR100 pKa = 11.84HH101 pKa = 5.94AVLARR106 pKa = 11.84HH107 pKa = 6.01AVLARR112 pKa = 11.84HH113 pKa = 6.01AVLARR118 pKa = 11.84HH119 pKa = 6.09AVPSRR124 pKa = 11.84HH125 pKa = 5.94AVLARR130 pKa = 11.84HH131 pKa = 6.15AVPARR136 pKa = 11.84HH137 pKa = 5.9AVPSRR142 pKa = 11.84RR143 pKa = 11.84AVLARR148 pKa = 11.84HH149 pKa = 5.87AVLARR154 pKa = 11.84HH155 pKa = 6.01AVLARR160 pKa = 11.84HH161 pKa = 6.01AVLARR166 pKa = 11.84HH167 pKa = 5.97AVLARR172 pKa = 11.84HH173 pKa = 5.56TVPARR178 pKa = 11.84HH179 pKa = 5.96AVLARR184 pKa = 11.84HH185 pKa = 6.01AVLARR190 pKa = 11.84HH191 pKa = 6.09AVPSRR196 pKa = 11.84HH197 pKa = 5.63AVPSRR202 pKa = 11.84RR203 pKa = 11.84AVLARR208 pKa = 11.84HH209 pKa = 5.87AVLARR214 pKa = 11.84HH215 pKa = 5.63TVPSRR220 pKa = 11.84HH221 pKa = 5.97AVLSLTSAIKK231 pKa = 10.23PSTARR236 pKa = 11.84AHH238 pKa = 6.41RR239 pKa = 11.84RR240 pKa = 11.84LPSAIASPTASAQNRR255 pKa = 11.84VSLSRR260 pKa = 11.84RR261 pKa = 11.84GFF263 pKa = 3.39
Molecular weight: 28.4 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1587121 |
30 |
3463 |
330.4 |
36.56 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.448 ± 0.051 | 0.766 ± 0.01 |
5.223 ± 0.024 | 6.388 ± 0.044 |
3.871 ± 0.025 | 7.589 ± 0.042 |
2.095 ± 0.019 | 6.274 ± 0.031 |
4.76 ± 0.035 | 9.8 ± 0.043 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.72 ± 0.017 | 3.721 ± 0.034 |
4.073 ± 0.023 | 4.009 ± 0.024 |
5.387 ± 0.042 | 6.435 ± 0.033 |
5.602 ± 0.045 | 7.153 ± 0.031 |
1.296 ± 0.015 | 3.386 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
