
Haloprofundus marisrubri
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Stenosarchaea group; Halobacteria; Haloferacales; Haloferacaceae; Haloprofundus
Average proteome isoelectric point is 5.02
Get precalculated fractions of proteins
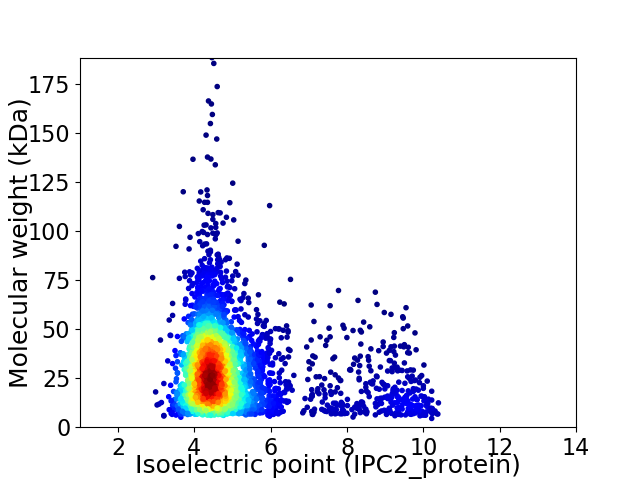
Virtual 2D-PAGE plot for 3761 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
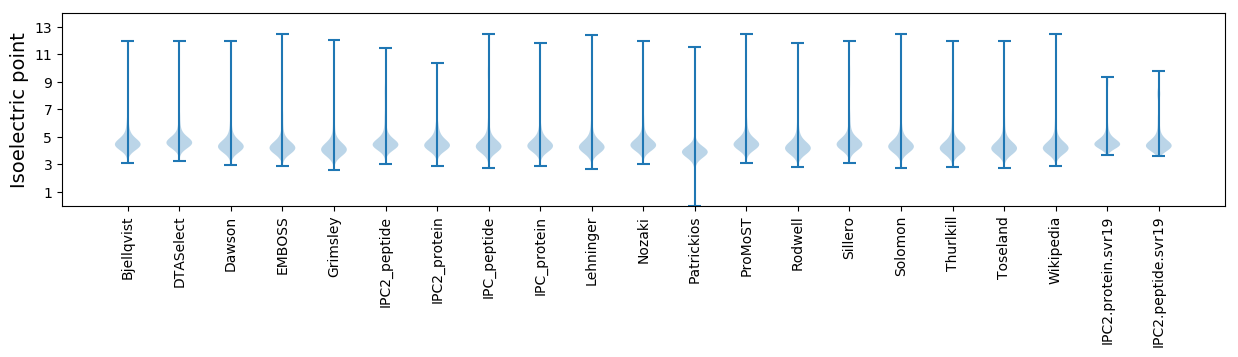
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0W1RD70|A0A0W1RD70_9EURY Peptidase M24 OS=Haloprofundus marisrubri OX=1514971 GN=AUR64_06345 PE=3 SV=1
MM1 pKa = 7.44KK2 pKa = 10.09FDD4 pKa = 3.72PSRR7 pKa = 11.84RR8 pKa = 11.84KK9 pKa = 9.96VLALGAGAALASTGLGSAADD29 pKa = 4.09LEE31 pKa = 4.79GAQNIDD37 pKa = 2.99SHH39 pKa = 6.16PTVDD43 pKa = 4.27AEE45 pKa = 4.61VVDD48 pKa = 4.21SFTDD52 pKa = 3.89GLEE55 pKa = 4.15LNVTISVTDD64 pKa = 3.87PDD66 pKa = 5.07DD67 pKa = 3.73EE68 pKa = 4.38VDD70 pKa = 3.84RR71 pKa = 11.84IGFIVEE77 pKa = 4.1NYY79 pKa = 10.07NDD81 pKa = 3.85RR82 pKa = 11.84TVDD85 pKa = 4.04IATHH89 pKa = 6.21WFNEE93 pKa = 3.74ASGEE97 pKa = 4.2KK98 pKa = 8.99TVSKK102 pKa = 10.66VVSLNRR108 pKa = 11.84APYY111 pKa = 9.58ACYY114 pKa = 10.5AFVKK118 pKa = 10.4AGSDD122 pKa = 4.03YY123 pKa = 11.26YY124 pKa = 11.47SVDD127 pKa = 3.33SVRR130 pKa = 11.84LGPEE134 pKa = 3.74TEE136 pKa = 4.36APAPQLLGVDD146 pKa = 3.91VSGTTATIDD155 pKa = 3.72YY156 pKa = 8.08EE157 pKa = 4.43VSPDD161 pKa = 3.3EE162 pKa = 4.07PAEE165 pKa = 4.05YY166 pKa = 10.47SVGASVSPISGSSSGDD182 pKa = 3.12PVYY185 pKa = 8.44TTEE188 pKa = 5.45RR189 pKa = 11.84NDD191 pKa = 3.87PYY193 pKa = 11.32DD194 pKa = 3.81EE195 pKa = 4.4PTTARR200 pKa = 11.84TVVTGLAPNTEE211 pKa = 4.36YY212 pKa = 10.23TATAWTEE219 pKa = 3.6LDD221 pKa = 2.9TWAMKK226 pKa = 9.8EE227 pKa = 3.76QSDD230 pKa = 4.25YY231 pKa = 11.88AEE233 pKa = 5.15SEE235 pKa = 4.22TQTFKK240 pKa = 10.76TGDD243 pKa = 3.43VVAPEE248 pKa = 4.59PEE250 pKa = 4.24TEE252 pKa = 3.8HH253 pKa = 7.3RR254 pKa = 11.84LVIGGSSDD262 pKa = 3.01VSYY265 pKa = 11.43YY266 pKa = 10.71RR267 pKa = 11.84VTTTGSLEE275 pKa = 4.11QTTEE279 pKa = 3.77TGDD282 pKa = 3.65APFSSATVDD291 pKa = 3.64DD292 pKa = 4.27EE293 pKa = 5.86DD294 pKa = 4.39YY295 pKa = 11.59VSGSSACGGVAGGADD310 pKa = 3.01AYY312 pKa = 11.4VFTGEE317 pKa = 4.13LTNVGVDD324 pKa = 4.02GAASVYY330 pKa = 10.81LDD332 pKa = 4.0GEE334 pKa = 4.56EE335 pKa = 5.03IDD337 pKa = 3.86PADD340 pKa = 3.82YY341 pKa = 11.19ADD343 pKa = 4.65EE344 pKa = 4.88LPHH347 pKa = 7.2HH348 pKa = 6.53LAITGGSDD356 pKa = 2.8LTEE359 pKa = 3.72YY360 pKa = 11.04AFRR363 pKa = 11.84VSGDD367 pKa = 3.58VIKK370 pKa = 8.46TTEE373 pKa = 3.66VDD375 pKa = 3.47GTGLSLNPSYY385 pKa = 11.2DD386 pKa = 3.91DD387 pKa = 4.14EE388 pKa = 4.76DD389 pKa = 4.24TVSAGQGAGATAGGTDD405 pKa = 3.86AYY407 pKa = 10.37RR408 pKa = 11.84FSSPQVTSAEE418 pKa = 3.99EE419 pKa = 4.82DD420 pKa = 3.1ISFTQFDD427 pKa = 3.88GDD429 pKa = 3.56ATVYY433 pKa = 11.11LNGEE437 pKa = 4.4EE438 pKa = 4.83IDD440 pKa = 3.9PADD443 pKa = 3.6YY444 pKa = 11.29
MM1 pKa = 7.44KK2 pKa = 10.09FDD4 pKa = 3.72PSRR7 pKa = 11.84RR8 pKa = 11.84KK9 pKa = 9.96VLALGAGAALASTGLGSAADD29 pKa = 4.09LEE31 pKa = 4.79GAQNIDD37 pKa = 2.99SHH39 pKa = 6.16PTVDD43 pKa = 4.27AEE45 pKa = 4.61VVDD48 pKa = 4.21SFTDD52 pKa = 3.89GLEE55 pKa = 4.15LNVTISVTDD64 pKa = 3.87PDD66 pKa = 5.07DD67 pKa = 3.73EE68 pKa = 4.38VDD70 pKa = 3.84RR71 pKa = 11.84IGFIVEE77 pKa = 4.1NYY79 pKa = 10.07NDD81 pKa = 3.85RR82 pKa = 11.84TVDD85 pKa = 4.04IATHH89 pKa = 6.21WFNEE93 pKa = 3.74ASGEE97 pKa = 4.2KK98 pKa = 8.99TVSKK102 pKa = 10.66VVSLNRR108 pKa = 11.84APYY111 pKa = 9.58ACYY114 pKa = 10.5AFVKK118 pKa = 10.4AGSDD122 pKa = 4.03YY123 pKa = 11.26YY124 pKa = 11.47SVDD127 pKa = 3.33SVRR130 pKa = 11.84LGPEE134 pKa = 3.74TEE136 pKa = 4.36APAPQLLGVDD146 pKa = 3.91VSGTTATIDD155 pKa = 3.72YY156 pKa = 8.08EE157 pKa = 4.43VSPDD161 pKa = 3.3EE162 pKa = 4.07PAEE165 pKa = 4.05YY166 pKa = 10.47SVGASVSPISGSSSGDD182 pKa = 3.12PVYY185 pKa = 8.44TTEE188 pKa = 5.45RR189 pKa = 11.84NDD191 pKa = 3.87PYY193 pKa = 11.32DD194 pKa = 3.81EE195 pKa = 4.4PTTARR200 pKa = 11.84TVVTGLAPNTEE211 pKa = 4.36YY212 pKa = 10.23TATAWTEE219 pKa = 3.6LDD221 pKa = 2.9TWAMKK226 pKa = 9.8EE227 pKa = 3.76QSDD230 pKa = 4.25YY231 pKa = 11.88AEE233 pKa = 5.15SEE235 pKa = 4.22TQTFKK240 pKa = 10.76TGDD243 pKa = 3.43VVAPEE248 pKa = 4.59PEE250 pKa = 4.24TEE252 pKa = 3.8HH253 pKa = 7.3RR254 pKa = 11.84LVIGGSSDD262 pKa = 3.01VSYY265 pKa = 11.43YY266 pKa = 10.71RR267 pKa = 11.84VTTTGSLEE275 pKa = 4.11QTTEE279 pKa = 3.77TGDD282 pKa = 3.65APFSSATVDD291 pKa = 3.64DD292 pKa = 4.27EE293 pKa = 5.86DD294 pKa = 4.39YY295 pKa = 11.59VSGSSACGGVAGGADD310 pKa = 3.01AYY312 pKa = 11.4VFTGEE317 pKa = 4.13LTNVGVDD324 pKa = 4.02GAASVYY330 pKa = 10.81LDD332 pKa = 4.0GEE334 pKa = 4.56EE335 pKa = 5.03IDD337 pKa = 3.86PADD340 pKa = 3.82YY341 pKa = 11.19ADD343 pKa = 4.65EE344 pKa = 4.88LPHH347 pKa = 7.2HH348 pKa = 6.53LAITGGSDD356 pKa = 2.8LTEE359 pKa = 3.72YY360 pKa = 11.04AFRR363 pKa = 11.84VSGDD367 pKa = 3.58VIKK370 pKa = 8.46TTEE373 pKa = 3.66VDD375 pKa = 3.47GTGLSLNPSYY385 pKa = 11.2DD386 pKa = 3.91DD387 pKa = 4.14EE388 pKa = 4.76DD389 pKa = 4.24TVSAGQGAGATAGGTDD405 pKa = 3.86AYY407 pKa = 10.37RR408 pKa = 11.84FSSPQVTSAEE418 pKa = 3.99EE419 pKa = 4.82DD420 pKa = 3.1ISFTQFDD427 pKa = 3.88GDD429 pKa = 3.56ATVYY433 pKa = 11.11LNGEE437 pKa = 4.4EE438 pKa = 4.83IDD440 pKa = 3.9PADD443 pKa = 3.6YY444 pKa = 11.29
Molecular weight: 46.63 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0W1REM5|A0A0W1REM5_9EURY Ferredoxin OS=Haloprofundus marisrubri OX=1514971 GN=AUR64_03090 PE=4 SV=1
MM1 pKa = 7.62SSGRR5 pKa = 11.84RR6 pKa = 11.84LRR8 pKa = 11.84IRR10 pKa = 11.84PEE12 pKa = 3.57PRR14 pKa = 11.84LEE16 pKa = 4.16TLHH19 pKa = 6.62EE20 pKa = 4.14RR21 pKa = 11.84AFKK24 pKa = 10.73YY25 pKa = 8.9ATLSIIGQNAPFLLAIEE42 pKa = 4.54PVRR45 pKa = 11.84EE46 pKa = 3.95SSPWGDD52 pKa = 3.28NPSNRR57 pKa = 11.84IHH59 pKa = 6.27RR60 pKa = 11.84VVRR63 pKa = 11.84RR64 pKa = 11.84LVRR67 pKa = 11.84RR68 pKa = 11.84AKK70 pKa = 9.82EE71 pKa = 3.68LVPIEE76 pKa = 4.14MVLCDD81 pKa = 4.62RR82 pKa = 11.84EE83 pKa = 4.31FDD85 pKa = 4.5SMDD88 pKa = 3.35AFQTLSNPNVNYY100 pKa = 10.26LIPKK104 pKa = 9.4RR105 pKa = 11.84VTSTEE110 pKa = 4.05STRR113 pKa = 11.84KK114 pKa = 9.66
MM1 pKa = 7.62SSGRR5 pKa = 11.84RR6 pKa = 11.84LRR8 pKa = 11.84IRR10 pKa = 11.84PEE12 pKa = 3.57PRR14 pKa = 11.84LEE16 pKa = 4.16TLHH19 pKa = 6.62EE20 pKa = 4.14RR21 pKa = 11.84AFKK24 pKa = 10.73YY25 pKa = 8.9ATLSIIGQNAPFLLAIEE42 pKa = 4.54PVRR45 pKa = 11.84EE46 pKa = 3.95SSPWGDD52 pKa = 3.28NPSNRR57 pKa = 11.84IHH59 pKa = 6.27RR60 pKa = 11.84VVRR63 pKa = 11.84RR64 pKa = 11.84LVRR67 pKa = 11.84RR68 pKa = 11.84AKK70 pKa = 9.82EE71 pKa = 3.68LVPIEE76 pKa = 4.14MVLCDD81 pKa = 4.62RR82 pKa = 11.84EE83 pKa = 4.31FDD85 pKa = 4.5SMDD88 pKa = 3.35AFQTLSNPNVNYY100 pKa = 10.26LIPKK104 pKa = 9.4RR105 pKa = 11.84VTSTEE110 pKa = 4.05STRR113 pKa = 11.84KK114 pKa = 9.66
Molecular weight: 13.24 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1074478 |
43 |
1681 |
285.7 |
31.11 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.431 ± 0.051 | 0.687 ± 0.013 |
7.996 ± 0.046 | 8.3 ± 0.051 |
3.554 ± 0.027 | 8.427 ± 0.045 |
1.978 ± 0.02 | 3.738 ± 0.033 |
2.0 ± 0.024 | 9.161 ± 0.055 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.774 ± 0.018 | 2.556 ± 0.024 |
4.493 ± 0.024 | 2.559 ± 0.024 |
6.557 ± 0.04 | 6.068 ± 0.038 |
6.427 ± 0.037 | 9.361 ± 0.042 |
1.147 ± 0.015 | 2.786 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
