
Raineya orbicola
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Cytophagia; Cytophagales; Raineyaceae; Raineya
Average proteome isoelectric point is 7.0
Get precalculated fractions of proteins
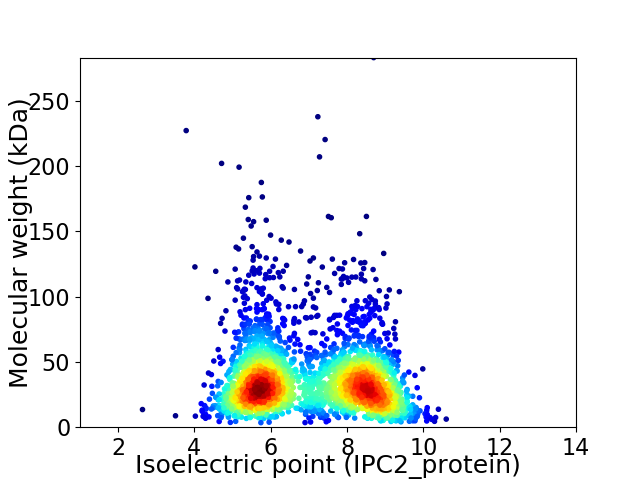
Virtual 2D-PAGE plot for 2684 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2N3IHX5|A0A2N3IHX5_9BACT Phosphoheptose isomerase OS=Raineya orbicola OX=2016530 GN=gmhA PE=3 SV=1
MM1 pKa = 7.54LSKK4 pKa = 9.64ITYY7 pKa = 9.09KK8 pKa = 10.8KK9 pKa = 10.79LFGFRR14 pKa = 11.84QGTGLTPNILGFGTILLLLYY34 pKa = 8.76WLNVTSSYY42 pKa = 11.21AQAGKK47 pKa = 10.38DD48 pKa = 3.23GALTVTTANTVLNRR62 pKa = 11.84YY63 pKa = 6.59TRR65 pKa = 11.84VTTDD69 pKa = 2.79VTAGSNVITVVNINDD84 pKa = 3.95LNRR87 pKa = 11.84DD88 pKa = 3.78GIGYY92 pKa = 9.57LPAGFVTNSSVYY104 pKa = 10.85ANNALSPGDD113 pKa = 4.61LIIIYY118 pKa = 8.58QAQGAIINSTNTINYY133 pKa = 8.06GQVTNYY139 pKa = 10.27NGAGTYY145 pKa = 9.77EE146 pKa = 4.35FAQVASVSGNNITLACRR163 pKa = 11.84TKK165 pKa = 10.42YY166 pKa = 10.7SYY168 pKa = 10.67FAARR172 pKa = 11.84YY173 pKa = 7.33VQVIRR178 pKa = 11.84VPQYY182 pKa = 7.4TTLTVNAGASVVAVPWGAPSFGGADD207 pKa = 3.43PSALEE212 pKa = 3.86RR213 pKa = 11.84RR214 pKa = 11.84RR215 pKa = 11.84GGFMVVHH222 pKa = 6.65ATNIVNNGTIHH233 pKa = 6.95ANAAGFRR240 pKa = 11.84GGTIDD245 pKa = 4.92PNTSPTGNNFSIAFVTNSSNEE266 pKa = 4.01SAEE269 pKa = 4.03KK270 pKa = 10.61GEE272 pKa = 4.58SIAGYY277 pKa = 10.21RR278 pKa = 11.84DD279 pKa = 4.41DD280 pKa = 5.87YY281 pKa = 12.03DD282 pKa = 3.31NLYY285 pKa = 9.84NGRR288 pKa = 11.84YY289 pKa = 9.28GRR291 pKa = 11.84GAPANGGGGGNGHH304 pKa = 6.34NAGGGGGANGGNPAGWFRR322 pKa = 11.84GAGVMNSFGTCGNPGAWALDD342 pKa = 3.42PDD344 pKa = 4.65YY345 pKa = 10.87IANGNALTNSQGGGRR360 pKa = 11.84GGYY363 pKa = 10.03SFGSSNQNACTLGPSYY379 pKa = 9.82PANFIAPGIPAADD392 pKa = 3.82VQNTAWGGDD401 pKa = 3.23NRR403 pKa = 11.84DD404 pKa = 3.41AVGGLGGRR412 pKa = 11.84PIVSANFQNQIFFGGGGGAGDD433 pKa = 4.25GNNGANADD441 pKa = 3.92GGDD444 pKa = 3.36GGGIVFLIATGNITGSGTISANGEE468 pKa = 3.89NGFNTISGHH477 pKa = 5.7NDD479 pKa = 2.78APGGGGGGGTVAIQATSIANTINIRR504 pKa = 11.84ANGGNGGNQLITNNEE519 pKa = 4.02SEE521 pKa = 4.54GPGGGGGGGVIVINATTDD539 pKa = 3.11GSTKK543 pKa = 10.06QVNGGQNGTTTSAAVTEE560 pKa = 5.09FTANGATSGNSGTISSVSVSLFSIVCTSNLGINKK594 pKa = 9.68AISNSSPNVGTNVTFTLTASNAGPDD619 pKa = 3.44PATNVVVNDD628 pKa = 4.82LLPAGYY634 pKa = 7.96TFVSATPSQGTYY646 pKa = 11.17NNVTGVWNVGSLGIGGSATLTIVATVNASGPYY678 pKa = 9.39TNTATISTPDD688 pKa = 3.28QNDD691 pKa = 3.63PNTSNNTSSVTPTPVPQTNLGVTKK715 pKa = 9.19TASNMTPPVGSNITFTITANNAGPSNATGVVVNDD749 pKa = 3.93VLPSGYY755 pKa = 8.66TFVSATTSQGTYY767 pKa = 10.52NNVSGVWSVGALANGASATLTITVTVNASGSYY799 pKa = 11.11ANTATITGDD808 pKa = 3.19QADD811 pKa = 4.07PVAGNNSSTATPTPVPQTNLGVTKK835 pKa = 9.19TASNMTPPVGSNITFTITANNAGPSNATGVVVNDD869 pKa = 3.93VLPSGYY875 pKa = 8.85TFVSATASQGTYY887 pKa = 10.82NNVSGVWSIGALANGASATLTITVTVNASGSYY919 pKa = 10.16TNTATISGDD928 pKa = 3.31QADD931 pKa = 4.22PVAGNNSSTATPTPVNVAPDD951 pKa = 3.94AVNDD955 pKa = 3.92NYY957 pKa = 11.61TMAQNGNLSNSVATNDD973 pKa = 3.04ILQAGGNPHH982 pKa = 6.88TFTLVSGGSAAANGTLTFNPDD1003 pKa = 2.54GTFTYY1008 pKa = 10.55VPNTGFSGTVSFTYY1022 pKa = 8.95QVCNGVTPINQCDD1035 pKa = 3.48TAVATIVVLPPPVAVNDD1052 pKa = 3.87NAVTPINTPVSGNLSTNDD1070 pKa = 3.55NLPSGGTYY1078 pKa = 9.58TYY1080 pKa = 10.55TIATPPNPAQGTVTINPDD1098 pKa = 2.67GTYY1101 pKa = 10.2TFTPATGFTGTATFTYY1117 pKa = 9.36QVCNQLGQCSTATVSVLVLPPPVAVNDD1144 pKa = 4.08TANTPINTPVNGNASTNDD1162 pKa = 3.48NLPSGGTYY1170 pKa = 9.65TYY1172 pKa = 10.9TVTTPPANGSVTMNPDD1188 pKa = 2.51GTYY1191 pKa = 9.93TYY1193 pKa = 10.29TPNTGFTGTDD1203 pKa = 2.69VFTYY1207 pKa = 7.97QVCNQVGQCSTATVTINVVPAPVANDD1233 pKa = 3.72DD1234 pKa = 4.73SYY1236 pKa = 12.18TMPQNGSLSNSVVGNDD1252 pKa = 3.35VLPASGNPHH1261 pKa = 5.85TFSLVSGGSAAANGTLTFNPDD1282 pKa = 2.54GTFTYY1287 pKa = 10.55VPNTGFSGTVSFTYY1301 pKa = 8.96QVCNSLIPAQCDD1313 pKa = 3.18TAVATIVVLPPPVAVNDD1330 pKa = 3.87NAVTPINTPVSGNLSTNDD1348 pKa = 3.55NLPSGGTYY1356 pKa = 9.58TYY1358 pKa = 10.55TIATPPNPAQGTVTINPDD1376 pKa = 2.67GTYY1379 pKa = 10.2TFTPATGFTGTATFTYY1395 pKa = 9.36QVCNQLGQCSTATVSVLVLPPPVAVNDD1422 pKa = 4.08TANTPINTPVNGNASTNDD1440 pKa = 3.48NLPSGGTYY1448 pKa = 9.65TYY1450 pKa = 10.9TVTTPPANGSVTMNPDD1466 pKa = 2.51GTYY1469 pKa = 9.93TYY1471 pKa = 10.29TPNTGFTGTDD1481 pKa = 2.69VFTYY1485 pKa = 7.97QVCNQVGQCSTATVTINVVPAPVANDD1511 pKa = 3.72DD1512 pKa = 4.73SYY1514 pKa = 12.18TMPQNGSLSNSVVGNDD1530 pKa = 3.35VLPASGNPHH1539 pKa = 5.85TFSLVSGGSAAANGTLTFNPDD1560 pKa = 2.54GTFTYY1565 pKa = 10.55VPNTGFSGTVSFTYY1579 pKa = 8.96QVCNSLIPAQCDD1591 pKa = 3.18TAVATIVVLPPPVAVNDD1608 pKa = 4.1SAVTPINTPVNGNLSSNDD1626 pKa = 3.4NLPAGGTYY1634 pKa = 9.57TYY1636 pKa = 10.64TVATPPNPAQGTVTINPDD1654 pKa = 2.67GTYY1657 pKa = 10.2TFTPATGFTGTASFTYY1673 pKa = 8.85QVCNQLGQCSTATVSVLVLPPPVAVNDD1700 pKa = 4.08TANTPINTPVNGNASTNDD1718 pKa = 3.48NLPSGGTYY1726 pKa = 9.65TYY1728 pKa = 10.9TVTTPPANGSVTMNPDD1744 pKa = 2.51GTYY1747 pKa = 9.93TYY1749 pKa = 10.29TPNTGFTGTDD1759 pKa = 2.69VFTYY1763 pKa = 7.97QVCNQVGQCSTATVTITVPANPVAVNDD1790 pKa = 4.22NYY1792 pKa = 8.04TTPINTPVGGNLAANDD1808 pKa = 4.23LLPTGFTYY1816 pKa = 10.27TFSLVNGGSAASNGTLVVNPDD1837 pKa = 2.74GTYY1840 pKa = 11.12NFTPNTGFSGVVSFTYY1856 pKa = 8.99QVCNQVNVCSTATVTITILAPPVASNDD1883 pKa = 3.79TAGTQPNTPVNGNVSPNDD1901 pKa = 3.43NLPAGGTYY1909 pKa = 9.72TFTVTTPPANGNVTMNPDD1927 pKa = 2.7GSYY1930 pKa = 10.06TYY1932 pKa = 9.88TPNPGFVGTDD1942 pKa = 2.82TFTYY1946 pKa = 8.07TVCNQLGQCSTATVTITVGQPPLAVNDD1973 pKa = 4.25EE1974 pKa = 4.58YY1975 pKa = 11.41NTGAGIPVSGNVGTNDD1991 pKa = 3.43QNPSGGTTTFNLVSGGTATQNGTLVFNPNGIFTFTPNPGFSGTVTFTYY2039 pKa = 9.0EE2040 pKa = 3.02ICNPVGCSQATVTINVAPPVQAVDD2064 pKa = 4.22DD2065 pKa = 4.82NYY2067 pKa = 8.9TTTPNNPVSGNLGTNDD2083 pKa = 3.5VVPAGQAPTFSLVNGGTATQNGTLVVNPDD2112 pKa = 2.9GSFTFTPNPGFVGTVTFTYY2131 pKa = 9.49QVCNQFGQCSTATVTIVIEE2150 pKa = 4.41DD2151 pKa = 3.92PVVQIPEE2158 pKa = 4.3GFSPNNDD2165 pKa = 3.8GINDD2169 pKa = 3.44NFVIKK2174 pKa = 10.24GRR2176 pKa = 11.84NGRR2179 pKa = 11.84PVVLRR2184 pKa = 11.84IYY2186 pKa = 10.41NRR2188 pKa = 11.84WGNLVYY2194 pKa = 10.91EE2195 pKa = 4.34NLNYY2199 pKa = 9.92QDD2201 pKa = 4.39DD2202 pKa = 4.2WNGTSNIGIRR2212 pKa = 11.84IGEE2215 pKa = 4.1QLPDD2219 pKa = 3.07GTYY2222 pKa = 10.3FYY2224 pKa = 10.29TAEE2227 pKa = 4.21FKK2229 pKa = 10.36DD2230 pKa = 3.16TGEE2233 pKa = 3.86RR2234 pKa = 11.84YY2235 pKa = 9.83ARR2237 pKa = 11.84YY2238 pKa = 8.43LTIQRR2243 pKa = 4.08
MM1 pKa = 7.54LSKK4 pKa = 9.64ITYY7 pKa = 9.09KK8 pKa = 10.8KK9 pKa = 10.79LFGFRR14 pKa = 11.84QGTGLTPNILGFGTILLLLYY34 pKa = 8.76WLNVTSSYY42 pKa = 11.21AQAGKK47 pKa = 10.38DD48 pKa = 3.23GALTVTTANTVLNRR62 pKa = 11.84YY63 pKa = 6.59TRR65 pKa = 11.84VTTDD69 pKa = 2.79VTAGSNVITVVNINDD84 pKa = 3.95LNRR87 pKa = 11.84DD88 pKa = 3.78GIGYY92 pKa = 9.57LPAGFVTNSSVYY104 pKa = 10.85ANNALSPGDD113 pKa = 4.61LIIIYY118 pKa = 8.58QAQGAIINSTNTINYY133 pKa = 8.06GQVTNYY139 pKa = 10.27NGAGTYY145 pKa = 9.77EE146 pKa = 4.35FAQVASVSGNNITLACRR163 pKa = 11.84TKK165 pKa = 10.42YY166 pKa = 10.7SYY168 pKa = 10.67FAARR172 pKa = 11.84YY173 pKa = 7.33VQVIRR178 pKa = 11.84VPQYY182 pKa = 7.4TTLTVNAGASVVAVPWGAPSFGGADD207 pKa = 3.43PSALEE212 pKa = 3.86RR213 pKa = 11.84RR214 pKa = 11.84RR215 pKa = 11.84GGFMVVHH222 pKa = 6.65ATNIVNNGTIHH233 pKa = 6.95ANAAGFRR240 pKa = 11.84GGTIDD245 pKa = 4.92PNTSPTGNNFSIAFVTNSSNEE266 pKa = 4.01SAEE269 pKa = 4.03KK270 pKa = 10.61GEE272 pKa = 4.58SIAGYY277 pKa = 10.21RR278 pKa = 11.84DD279 pKa = 4.41DD280 pKa = 5.87YY281 pKa = 12.03DD282 pKa = 3.31NLYY285 pKa = 9.84NGRR288 pKa = 11.84YY289 pKa = 9.28GRR291 pKa = 11.84GAPANGGGGGNGHH304 pKa = 6.34NAGGGGGANGGNPAGWFRR322 pKa = 11.84GAGVMNSFGTCGNPGAWALDD342 pKa = 3.42PDD344 pKa = 4.65YY345 pKa = 10.87IANGNALTNSQGGGRR360 pKa = 11.84GGYY363 pKa = 10.03SFGSSNQNACTLGPSYY379 pKa = 9.82PANFIAPGIPAADD392 pKa = 3.82VQNTAWGGDD401 pKa = 3.23NRR403 pKa = 11.84DD404 pKa = 3.41AVGGLGGRR412 pKa = 11.84PIVSANFQNQIFFGGGGGAGDD433 pKa = 4.25GNNGANADD441 pKa = 3.92GGDD444 pKa = 3.36GGGIVFLIATGNITGSGTISANGEE468 pKa = 3.89NGFNTISGHH477 pKa = 5.7NDD479 pKa = 2.78APGGGGGGGTVAIQATSIANTINIRR504 pKa = 11.84ANGGNGGNQLITNNEE519 pKa = 4.02SEE521 pKa = 4.54GPGGGGGGGVIVINATTDD539 pKa = 3.11GSTKK543 pKa = 10.06QVNGGQNGTTTSAAVTEE560 pKa = 5.09FTANGATSGNSGTISSVSVSLFSIVCTSNLGINKK594 pKa = 9.68AISNSSPNVGTNVTFTLTASNAGPDD619 pKa = 3.44PATNVVVNDD628 pKa = 4.82LLPAGYY634 pKa = 7.96TFVSATPSQGTYY646 pKa = 11.17NNVTGVWNVGSLGIGGSATLTIVATVNASGPYY678 pKa = 9.39TNTATISTPDD688 pKa = 3.28QNDD691 pKa = 3.63PNTSNNTSSVTPTPVPQTNLGVTKK715 pKa = 9.19TASNMTPPVGSNITFTITANNAGPSNATGVVVNDD749 pKa = 3.93VLPSGYY755 pKa = 8.66TFVSATTSQGTYY767 pKa = 10.52NNVSGVWSVGALANGASATLTITVTVNASGSYY799 pKa = 11.11ANTATITGDD808 pKa = 3.19QADD811 pKa = 4.07PVAGNNSSTATPTPVPQTNLGVTKK835 pKa = 9.19TASNMTPPVGSNITFTITANNAGPSNATGVVVNDD869 pKa = 3.93VLPSGYY875 pKa = 8.85TFVSATASQGTYY887 pKa = 10.82NNVSGVWSIGALANGASATLTITVTVNASGSYY919 pKa = 10.16TNTATISGDD928 pKa = 3.31QADD931 pKa = 4.22PVAGNNSSTATPTPVNVAPDD951 pKa = 3.94AVNDD955 pKa = 3.92NYY957 pKa = 11.61TMAQNGNLSNSVATNDD973 pKa = 3.04ILQAGGNPHH982 pKa = 6.88TFTLVSGGSAAANGTLTFNPDD1003 pKa = 2.54GTFTYY1008 pKa = 10.55VPNTGFSGTVSFTYY1022 pKa = 8.95QVCNGVTPINQCDD1035 pKa = 3.48TAVATIVVLPPPVAVNDD1052 pKa = 3.87NAVTPINTPVSGNLSTNDD1070 pKa = 3.55NLPSGGTYY1078 pKa = 9.58TYY1080 pKa = 10.55TIATPPNPAQGTVTINPDD1098 pKa = 2.67GTYY1101 pKa = 10.2TFTPATGFTGTATFTYY1117 pKa = 9.36QVCNQLGQCSTATVSVLVLPPPVAVNDD1144 pKa = 4.08TANTPINTPVNGNASTNDD1162 pKa = 3.48NLPSGGTYY1170 pKa = 9.65TYY1172 pKa = 10.9TVTTPPANGSVTMNPDD1188 pKa = 2.51GTYY1191 pKa = 9.93TYY1193 pKa = 10.29TPNTGFTGTDD1203 pKa = 2.69VFTYY1207 pKa = 7.97QVCNQVGQCSTATVTINVVPAPVANDD1233 pKa = 3.72DD1234 pKa = 4.73SYY1236 pKa = 12.18TMPQNGSLSNSVVGNDD1252 pKa = 3.35VLPASGNPHH1261 pKa = 5.85TFSLVSGGSAAANGTLTFNPDD1282 pKa = 2.54GTFTYY1287 pKa = 10.55VPNTGFSGTVSFTYY1301 pKa = 8.96QVCNSLIPAQCDD1313 pKa = 3.18TAVATIVVLPPPVAVNDD1330 pKa = 3.87NAVTPINTPVSGNLSTNDD1348 pKa = 3.55NLPSGGTYY1356 pKa = 9.58TYY1358 pKa = 10.55TIATPPNPAQGTVTINPDD1376 pKa = 2.67GTYY1379 pKa = 10.2TFTPATGFTGTATFTYY1395 pKa = 9.36QVCNQLGQCSTATVSVLVLPPPVAVNDD1422 pKa = 4.08TANTPINTPVNGNASTNDD1440 pKa = 3.48NLPSGGTYY1448 pKa = 9.65TYY1450 pKa = 10.9TVTTPPANGSVTMNPDD1466 pKa = 2.51GTYY1469 pKa = 9.93TYY1471 pKa = 10.29TPNTGFTGTDD1481 pKa = 2.69VFTYY1485 pKa = 7.97QVCNQVGQCSTATVTINVVPAPVANDD1511 pKa = 3.72DD1512 pKa = 4.73SYY1514 pKa = 12.18TMPQNGSLSNSVVGNDD1530 pKa = 3.35VLPASGNPHH1539 pKa = 5.85TFSLVSGGSAAANGTLTFNPDD1560 pKa = 2.54GTFTYY1565 pKa = 10.55VPNTGFSGTVSFTYY1579 pKa = 8.96QVCNSLIPAQCDD1591 pKa = 3.18TAVATIVVLPPPVAVNDD1608 pKa = 4.1SAVTPINTPVNGNLSSNDD1626 pKa = 3.4NLPAGGTYY1634 pKa = 9.57TYY1636 pKa = 10.64TVATPPNPAQGTVTINPDD1654 pKa = 2.67GTYY1657 pKa = 10.2TFTPATGFTGTASFTYY1673 pKa = 8.85QVCNQLGQCSTATVSVLVLPPPVAVNDD1700 pKa = 4.08TANTPINTPVNGNASTNDD1718 pKa = 3.48NLPSGGTYY1726 pKa = 9.65TYY1728 pKa = 10.9TVTTPPANGSVTMNPDD1744 pKa = 2.51GTYY1747 pKa = 9.93TYY1749 pKa = 10.29TPNTGFTGTDD1759 pKa = 2.69VFTYY1763 pKa = 7.97QVCNQVGQCSTATVTITVPANPVAVNDD1790 pKa = 4.22NYY1792 pKa = 8.04TTPINTPVGGNLAANDD1808 pKa = 4.23LLPTGFTYY1816 pKa = 10.27TFSLVNGGSAASNGTLVVNPDD1837 pKa = 2.74GTYY1840 pKa = 11.12NFTPNTGFSGVVSFTYY1856 pKa = 8.99QVCNQVNVCSTATVTITILAPPVASNDD1883 pKa = 3.79TAGTQPNTPVNGNVSPNDD1901 pKa = 3.43NLPAGGTYY1909 pKa = 9.72TFTVTTPPANGNVTMNPDD1927 pKa = 2.7GSYY1930 pKa = 10.06TYY1932 pKa = 9.88TPNPGFVGTDD1942 pKa = 2.82TFTYY1946 pKa = 8.07TVCNQLGQCSTATVTITVGQPPLAVNDD1973 pKa = 4.25EE1974 pKa = 4.58YY1975 pKa = 11.41NTGAGIPVSGNVGTNDD1991 pKa = 3.43QNPSGGTTTFNLVSGGTATQNGTLVFNPNGIFTFTPNPGFSGTVTFTYY2039 pKa = 9.0EE2040 pKa = 3.02ICNPVGCSQATVTINVAPPVQAVDD2064 pKa = 4.22DD2065 pKa = 4.82NYY2067 pKa = 8.9TTTPNNPVSGNLGTNDD2083 pKa = 3.5VVPAGQAPTFSLVNGGTATQNGTLVVNPDD2112 pKa = 2.9GSFTFTPNPGFVGTVTFTYY2131 pKa = 9.49QVCNQFGQCSTATVTIVIEE2150 pKa = 4.41DD2151 pKa = 3.92PVVQIPEE2158 pKa = 4.3GFSPNNDD2165 pKa = 3.8GINDD2169 pKa = 3.44NFVIKK2174 pKa = 10.24GRR2176 pKa = 11.84NGRR2179 pKa = 11.84PVVLRR2184 pKa = 11.84IYY2186 pKa = 10.41NRR2188 pKa = 11.84WGNLVYY2194 pKa = 10.91EE2195 pKa = 4.34NLNYY2199 pKa = 9.92QDD2201 pKa = 4.39DD2202 pKa = 4.2WNGTSNIGIRR2212 pKa = 11.84IGEE2215 pKa = 4.1QLPDD2219 pKa = 3.07GTYY2222 pKa = 10.3FYY2224 pKa = 10.29TAEE2227 pKa = 4.21FKK2229 pKa = 10.36DD2230 pKa = 3.16TGEE2233 pKa = 3.86RR2234 pKa = 11.84YY2235 pKa = 9.83ARR2237 pKa = 11.84YY2238 pKa = 8.43LTIQRR2243 pKa = 4.08
Molecular weight: 227.31 kDa
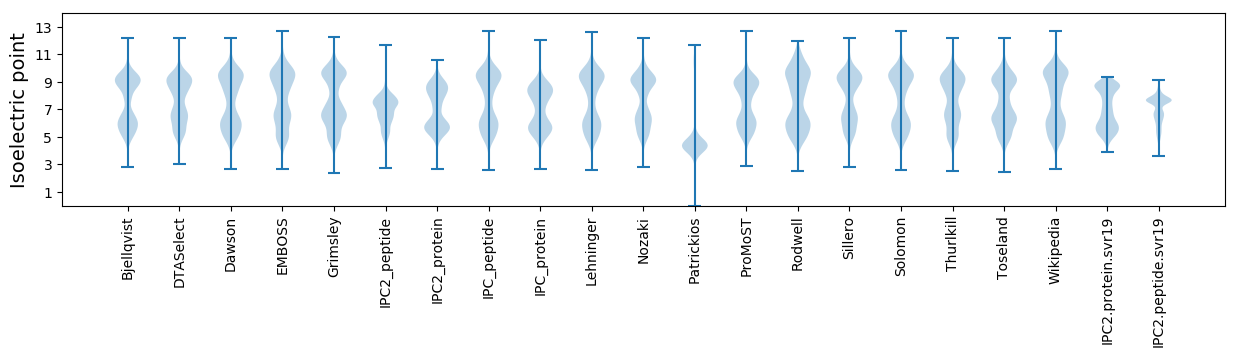
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2N3I946|A0A2N3I946_9BACT Outer membrane protein beta-barrel domain OS=Raineya orbicola OX=2016530 GN=Rain11_2252 PE=4 SV=1
MM1 pKa = 7.71IYY3 pKa = 10.38SLHH6 pKa = 6.37KK7 pKa = 10.15RR8 pKa = 11.84GASRR12 pKa = 11.84IVRR15 pKa = 11.84PFLVANYY22 pKa = 10.01YY23 pKa = 9.03NVKK26 pKa = 10.83GNFATFNWLLLVLALLILFASSVAVGSVSIPFTEE60 pKa = 4.04IVKK63 pKa = 9.06TLSGLQEE70 pKa = 3.65NATWKK75 pKa = 10.4FIVWEE80 pKa = 3.99LRR82 pKa = 11.84LPKK85 pKa = 10.41ALTAVLVGVALSVAGLVMQTFFANPLASPSEE116 pKa = 4.25LGVSAGASLGVASITLSSGISWSAFQQWGLVGNSFVAIIAILGAMLVMLLLLLMSRR172 pKa = 11.84FVWNNTTLLIVGLMLGSLAISLIGLWQFLSSPEE205 pKa = 4.0QIRR208 pKa = 11.84DD209 pKa = 3.74FLWWSFGSLAGTNYY223 pKa = 10.5SQIAVLAFLVAMSLLLLFLQTQKK246 pKa = 11.34LNALLLGEE254 pKa = 4.48MYY256 pKa = 10.61ARR258 pKa = 11.84SMGVNIRR265 pKa = 11.84KK266 pKa = 8.91LRR268 pKa = 11.84WVLIVVVSVLSGVTTAFCGPIGFVGLAVPHH298 pKa = 6.8LARR301 pKa = 11.84SLFQTANHH309 pKa = 5.64WTLFPASTLLGASVLLFCDD328 pKa = 3.2ILAYY332 pKa = 10.99NLFANFALPINLVTSLLGSPIVIWIVIRR360 pKa = 11.84RR361 pKa = 11.84SFRR364 pKa = 11.84FF365 pKa = 3.22
MM1 pKa = 7.71IYY3 pKa = 10.38SLHH6 pKa = 6.37KK7 pKa = 10.15RR8 pKa = 11.84GASRR12 pKa = 11.84IVRR15 pKa = 11.84PFLVANYY22 pKa = 10.01YY23 pKa = 9.03NVKK26 pKa = 10.83GNFATFNWLLLVLALLILFASSVAVGSVSIPFTEE60 pKa = 4.04IVKK63 pKa = 9.06TLSGLQEE70 pKa = 3.65NATWKK75 pKa = 10.4FIVWEE80 pKa = 3.99LRR82 pKa = 11.84LPKK85 pKa = 10.41ALTAVLVGVALSVAGLVMQTFFANPLASPSEE116 pKa = 4.25LGVSAGASLGVASITLSSGISWSAFQQWGLVGNSFVAIIAILGAMLVMLLLLLMSRR172 pKa = 11.84FVWNNTTLLIVGLMLGSLAISLIGLWQFLSSPEE205 pKa = 4.0QIRR208 pKa = 11.84DD209 pKa = 3.74FLWWSFGSLAGTNYY223 pKa = 10.5SQIAVLAFLVAMSLLLLFLQTQKK246 pKa = 11.34LNALLLGEE254 pKa = 4.48MYY256 pKa = 10.61ARR258 pKa = 11.84SMGVNIRR265 pKa = 11.84KK266 pKa = 8.91LRR268 pKa = 11.84WVLIVVVSVLSGVTTAFCGPIGFVGLAVPHH298 pKa = 6.8LARR301 pKa = 11.84SLFQTANHH309 pKa = 5.64WTLFPASTLLGASVLLFCDD328 pKa = 3.2ILAYY332 pKa = 10.99NLFANFALPINLVTSLLGSPIVIWIVIRR360 pKa = 11.84RR361 pKa = 11.84SFRR364 pKa = 11.84FF365 pKa = 3.22
Molecular weight: 39.69 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
933702 |
29 |
2470 |
347.9 |
39.6 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.918 ± 0.044 | 0.854 ± 0.014 |
4.418 ± 0.027 | 7.171 ± 0.063 |
5.538 ± 0.04 | 5.914 ± 0.051 |
1.727 ± 0.021 | 7.718 ± 0.046 |
7.903 ± 0.064 | 9.86 ± 0.056 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.057 ± 0.019 | 5.588 ± 0.046 |
3.644 ± 0.031 | 4.618 ± 0.031 |
3.968 ± 0.031 | 5.655 ± 0.027 |
5.249 ± 0.048 | 5.915 ± 0.041 |
1.229 ± 0.019 | 4.057 ± 0.028 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
