
Hubei yanvirus-like virus 1
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.09
Get precalculated fractions of proteins
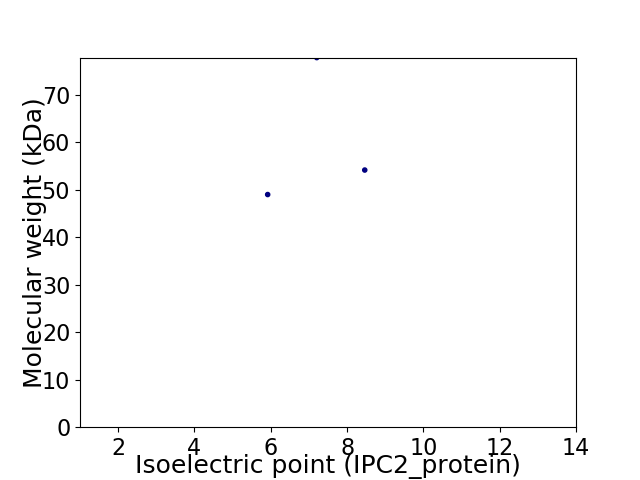
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
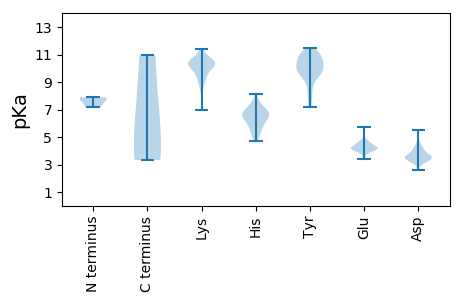
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A1L3KLE7|A0A1L3KLE7_9VIRU RNA-dependent RNA polymerase OS=Hubei yanvirus-like virus 1 OX=1923343 PE=4 SV=1
MM1 pKa = 7.19QVWEE5 pKa = 5.43WICTMPSRR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84KK16 pKa = 8.94QLVRR20 pKa = 11.84AFKK23 pKa = 10.62ARR25 pKa = 11.84EE26 pKa = 3.86EE27 pKa = 4.19LGEE30 pKa = 4.51DD31 pKa = 3.32SPEE34 pKa = 3.9YY35 pKa = 10.34EE36 pKa = 4.15IIKK39 pKa = 10.57AFVKK43 pKa = 10.28TEE45 pKa = 3.78LLPYY49 pKa = 10.37FGVADD54 pKa = 3.57GDD56 pKa = 3.89YY57 pKa = 11.19SVFQVEE63 pKa = 4.12YY64 pKa = 9.7VARR67 pKa = 11.84LIQAPHH73 pKa = 7.32DD74 pKa = 4.05EE75 pKa = 4.44THH77 pKa = 6.99LDD79 pKa = 3.22AGPYY83 pKa = 9.67LKK85 pKa = 10.59PLVQRR90 pKa = 11.84LKK92 pKa = 9.88EE93 pKa = 4.15TWSWEE98 pKa = 3.38HH99 pKa = 5.24WLFYY103 pKa = 11.04ASVSPEE109 pKa = 4.79KK110 pKa = 10.45LDD112 pKa = 3.09KK113 pKa = 10.21WLNKK117 pKa = 9.58HH118 pKa = 6.43RR119 pKa = 11.84GATSWFWSDD128 pKa = 3.42YY129 pKa = 11.09SSFDD133 pKa = 3.32STYY136 pKa = 10.97SAHH139 pKa = 5.11TWRR142 pKa = 11.84MIEE145 pKa = 4.23RR146 pKa = 11.84FYY148 pKa = 10.65QQIYY152 pKa = 9.72PEE154 pKa = 4.56APEE157 pKa = 4.5SLWKK161 pKa = 10.53ALNAWRR167 pKa = 11.84KK168 pKa = 7.37PHH170 pKa = 5.82GKK172 pKa = 9.27VRR174 pKa = 11.84SHH176 pKa = 7.22RR177 pKa = 11.84DD178 pKa = 3.42DD179 pKa = 4.45AKK181 pKa = 11.16LEE183 pKa = 4.02YY184 pKa = 9.84FAPIMNASGRR194 pKa = 11.84DD195 pKa = 3.49DD196 pKa = 3.48TALANALVNGIVLALSFTAVLAGKK220 pKa = 9.96SIEE223 pKa = 4.28SLEE226 pKa = 4.16EE227 pKa = 4.78ADD229 pKa = 4.11VLWASRR235 pKa = 11.84QFSIAVVGDD244 pKa = 4.01DD245 pKa = 4.54SLVACSFDD253 pKa = 3.34VRR255 pKa = 11.84PLVGAIEE262 pKa = 4.21GKK264 pKa = 9.39IRR266 pKa = 11.84RR267 pKa = 11.84FGLVAKK273 pKa = 9.64CFSSEE278 pKa = 4.13EE279 pKa = 4.19LCDD282 pKa = 3.67VTFLGMMPYY291 pKa = 9.95PVAGDD296 pKa = 4.44LYY298 pKa = 9.12WGPTIGRR305 pKa = 11.84RR306 pKa = 11.84LYY308 pKa = 10.59KK309 pKa = 10.49AFWQADD315 pKa = 3.66PKK317 pKa = 11.4GNLPAWTLGVAKK329 pKa = 10.27QLALYY334 pKa = 9.84QCVPVLSEE342 pKa = 3.49IANRR346 pKa = 11.84VCTLLDD352 pKa = 3.5GGKK355 pKa = 8.19ITRR358 pKa = 11.84QAVDD362 pKa = 3.46EE363 pKa = 4.14NRR365 pKa = 11.84VWSARR370 pKa = 11.84SSATPPYY377 pKa = 10.27DD378 pKa = 3.49YY379 pKa = 11.03STMVWMCKK387 pKa = 8.91RR388 pKa = 11.84YY389 pKa = 9.67RR390 pKa = 11.84EE391 pKa = 4.08QGLVPRR397 pKa = 11.84AMAADD402 pKa = 3.72VEE404 pKa = 4.72TVEE407 pKa = 5.72RR408 pKa = 11.84IDD410 pKa = 3.59RR411 pKa = 11.84LPAIVRR417 pKa = 11.84LLTVDD422 pKa = 3.87ASLLVDD428 pKa = 4.52DD429 pKa = 5.53LL430 pKa = 4.71
MM1 pKa = 7.19QVWEE5 pKa = 5.43WICTMPSRR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84KK16 pKa = 8.94QLVRR20 pKa = 11.84AFKK23 pKa = 10.62ARR25 pKa = 11.84EE26 pKa = 3.86EE27 pKa = 4.19LGEE30 pKa = 4.51DD31 pKa = 3.32SPEE34 pKa = 3.9YY35 pKa = 10.34EE36 pKa = 4.15IIKK39 pKa = 10.57AFVKK43 pKa = 10.28TEE45 pKa = 3.78LLPYY49 pKa = 10.37FGVADD54 pKa = 3.57GDD56 pKa = 3.89YY57 pKa = 11.19SVFQVEE63 pKa = 4.12YY64 pKa = 9.7VARR67 pKa = 11.84LIQAPHH73 pKa = 7.32DD74 pKa = 4.05EE75 pKa = 4.44THH77 pKa = 6.99LDD79 pKa = 3.22AGPYY83 pKa = 9.67LKK85 pKa = 10.59PLVQRR90 pKa = 11.84LKK92 pKa = 9.88EE93 pKa = 4.15TWSWEE98 pKa = 3.38HH99 pKa = 5.24WLFYY103 pKa = 11.04ASVSPEE109 pKa = 4.79KK110 pKa = 10.45LDD112 pKa = 3.09KK113 pKa = 10.21WLNKK117 pKa = 9.58HH118 pKa = 6.43RR119 pKa = 11.84GATSWFWSDD128 pKa = 3.42YY129 pKa = 11.09SSFDD133 pKa = 3.32STYY136 pKa = 10.97SAHH139 pKa = 5.11TWRR142 pKa = 11.84MIEE145 pKa = 4.23RR146 pKa = 11.84FYY148 pKa = 10.65QQIYY152 pKa = 9.72PEE154 pKa = 4.56APEE157 pKa = 4.5SLWKK161 pKa = 10.53ALNAWRR167 pKa = 11.84KK168 pKa = 7.37PHH170 pKa = 5.82GKK172 pKa = 9.27VRR174 pKa = 11.84SHH176 pKa = 7.22RR177 pKa = 11.84DD178 pKa = 3.42DD179 pKa = 4.45AKK181 pKa = 11.16LEE183 pKa = 4.02YY184 pKa = 9.84FAPIMNASGRR194 pKa = 11.84DD195 pKa = 3.49DD196 pKa = 3.48TALANALVNGIVLALSFTAVLAGKK220 pKa = 9.96SIEE223 pKa = 4.28SLEE226 pKa = 4.16EE227 pKa = 4.78ADD229 pKa = 4.11VLWASRR235 pKa = 11.84QFSIAVVGDD244 pKa = 4.01DD245 pKa = 4.54SLVACSFDD253 pKa = 3.34VRR255 pKa = 11.84PLVGAIEE262 pKa = 4.21GKK264 pKa = 9.39IRR266 pKa = 11.84RR267 pKa = 11.84FGLVAKK273 pKa = 9.64CFSSEE278 pKa = 4.13EE279 pKa = 4.19LCDD282 pKa = 3.67VTFLGMMPYY291 pKa = 9.95PVAGDD296 pKa = 4.44LYY298 pKa = 9.12WGPTIGRR305 pKa = 11.84RR306 pKa = 11.84LYY308 pKa = 10.59KK309 pKa = 10.49AFWQADD315 pKa = 3.66PKK317 pKa = 11.4GNLPAWTLGVAKK329 pKa = 10.27QLALYY334 pKa = 9.84QCVPVLSEE342 pKa = 3.49IANRR346 pKa = 11.84VCTLLDD352 pKa = 3.5GGKK355 pKa = 8.19ITRR358 pKa = 11.84QAVDD362 pKa = 3.46EE363 pKa = 4.14NRR365 pKa = 11.84VWSARR370 pKa = 11.84SSATPPYY377 pKa = 10.27DD378 pKa = 3.49YY379 pKa = 11.03STMVWMCKK387 pKa = 8.91RR388 pKa = 11.84YY389 pKa = 9.67RR390 pKa = 11.84EE391 pKa = 4.08QGLVPRR397 pKa = 11.84AMAADD402 pKa = 3.72VEE404 pKa = 4.72TVEE407 pKa = 5.72RR408 pKa = 11.84IDD410 pKa = 3.59RR411 pKa = 11.84LPAIVRR417 pKa = 11.84LLTVDD422 pKa = 3.87ASLLVDD428 pKa = 4.52DD429 pKa = 5.53LL430 pKa = 4.71
Molecular weight: 49.03 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KLE7|A0A1L3KLE7_9VIRU RNA-dependent RNA polymerase OS=Hubei yanvirus-like virus 1 OX=1923343 PE=4 SV=1
MM1 pKa = 7.8ISEE4 pKa = 4.6PFWLASLCFSTTTAEE19 pKa = 4.08MPLTKK24 pKa = 10.15VRR26 pKa = 11.84NATSGVSDD34 pKa = 3.47IAKK37 pKa = 10.32AIAIPQEE44 pKa = 3.91HH45 pKa = 6.43RR46 pKa = 11.84PIRR49 pKa = 11.84IPTYY53 pKa = 9.77PSLEE57 pKa = 4.02RR58 pKa = 11.84SAVLHH63 pKa = 5.72LQQTRR68 pKa = 11.84TLKK71 pKa = 10.78VGAAGYY77 pKa = 9.89FDD79 pKa = 5.32AMLFRR84 pKa = 11.84SPVHH88 pKa = 5.99PLWGTYY94 pKa = 8.28QPSTSNAAVYY104 pKa = 9.79CSWNYY109 pKa = 7.79TASTILTTTTSMPINAGVFGTPQGVGVGGPDD140 pKa = 3.08WAAVTPVGYY149 pKa = 10.63GKK151 pKa = 10.34DD152 pKa = 3.75GLPYY156 pKa = 9.46MYY158 pKa = 10.21IPANVLFQATLNSTATFTTWEE179 pKa = 4.24MDD181 pKa = 3.46LEE183 pKa = 4.65CYY185 pKa = 10.62YY186 pKa = 10.66GTDD189 pKa = 3.59DD190 pKa = 3.62SSEE193 pKa = 4.15RR194 pKa = 11.84LSIMLTGSGVFGNGLSINANAWVRR218 pKa = 11.84PVALRR223 pKa = 11.84FTAAPTVAGNLTYY236 pKa = 11.31VNIGFFGGGSFNAPNAPIVASFFPLFPVPEE266 pKa = 4.26IQNSVTPYY274 pKa = 9.99YY275 pKa = 9.64STRR278 pKa = 11.84CTAAAVLFSNVTAVLQKK295 pKa = 10.68EE296 pKa = 4.6GTVQAGRR303 pKa = 11.84LNVSSGYY310 pKa = 9.91PYY312 pKa = 10.88DD313 pKa = 3.79PSFSFGVLTSLHH325 pKa = 5.73PQEE328 pKa = 5.36KK329 pKa = 10.11YY330 pKa = 10.15FGKK333 pKa = 10.86LEE335 pKa = 4.69DD336 pKa = 3.57GLYY339 pKa = 10.84AFSLPSAEE347 pKa = 3.9SDD349 pKa = 3.5TFRR352 pKa = 11.84DD353 pKa = 4.26CVSLSAIASKK363 pKa = 10.25PFPLFHH369 pKa = 7.53IEE371 pKa = 3.88GFGYY375 pKa = 7.98YY376 pKa = 7.75QHH378 pKa = 6.49MRR380 pKa = 11.84FSDD383 pKa = 4.1LDD385 pKa = 3.63ATSATQLAITLDD397 pKa = 3.4YY398 pKa = 10.72HH399 pKa = 8.15LEE401 pKa = 4.04FRR403 pKa = 11.84SSSMLFNLGYY413 pKa = 10.55SSLPLEE419 pKa = 5.07AYY421 pKa = 10.39HH422 pKa = 6.37EE423 pKa = 4.43AQLALVKK430 pKa = 10.29MGTFFEE436 pKa = 4.47NPKK439 pKa = 10.19HH440 pKa = 6.38ISQITKK446 pKa = 10.06KK447 pKa = 10.56VLNSLPKK454 pKa = 9.41VRR456 pKa = 11.84APALVVMKK464 pKa = 8.43VTKK467 pKa = 10.47GAGRR471 pKa = 11.84PDD473 pKa = 3.51GSKK476 pKa = 10.73GGKK479 pKa = 8.94KK480 pKa = 10.3DD481 pKa = 3.65NAPKK485 pKa = 10.49QNGGKK490 pKa = 9.92GKK492 pKa = 10.18KK493 pKa = 9.13SKK495 pKa = 10.24KK496 pKa = 10.52GKK498 pKa = 9.75DD499 pKa = 3.06KK500 pKa = 11.03KK501 pKa = 10.95
MM1 pKa = 7.8ISEE4 pKa = 4.6PFWLASLCFSTTTAEE19 pKa = 4.08MPLTKK24 pKa = 10.15VRR26 pKa = 11.84NATSGVSDD34 pKa = 3.47IAKK37 pKa = 10.32AIAIPQEE44 pKa = 3.91HH45 pKa = 6.43RR46 pKa = 11.84PIRR49 pKa = 11.84IPTYY53 pKa = 9.77PSLEE57 pKa = 4.02RR58 pKa = 11.84SAVLHH63 pKa = 5.72LQQTRR68 pKa = 11.84TLKK71 pKa = 10.78VGAAGYY77 pKa = 9.89FDD79 pKa = 5.32AMLFRR84 pKa = 11.84SPVHH88 pKa = 5.99PLWGTYY94 pKa = 8.28QPSTSNAAVYY104 pKa = 9.79CSWNYY109 pKa = 7.79TASTILTTTTSMPINAGVFGTPQGVGVGGPDD140 pKa = 3.08WAAVTPVGYY149 pKa = 10.63GKK151 pKa = 10.34DD152 pKa = 3.75GLPYY156 pKa = 9.46MYY158 pKa = 10.21IPANVLFQATLNSTATFTTWEE179 pKa = 4.24MDD181 pKa = 3.46LEE183 pKa = 4.65CYY185 pKa = 10.62YY186 pKa = 10.66GTDD189 pKa = 3.59DD190 pKa = 3.62SSEE193 pKa = 4.15RR194 pKa = 11.84LSIMLTGSGVFGNGLSINANAWVRR218 pKa = 11.84PVALRR223 pKa = 11.84FTAAPTVAGNLTYY236 pKa = 11.31VNIGFFGGGSFNAPNAPIVASFFPLFPVPEE266 pKa = 4.26IQNSVTPYY274 pKa = 9.99YY275 pKa = 9.64STRR278 pKa = 11.84CTAAAVLFSNVTAVLQKK295 pKa = 10.68EE296 pKa = 4.6GTVQAGRR303 pKa = 11.84LNVSSGYY310 pKa = 9.91PYY312 pKa = 10.88DD313 pKa = 3.79PSFSFGVLTSLHH325 pKa = 5.73PQEE328 pKa = 5.36KK329 pKa = 10.11YY330 pKa = 10.15FGKK333 pKa = 10.86LEE335 pKa = 4.69DD336 pKa = 3.57GLYY339 pKa = 10.84AFSLPSAEE347 pKa = 3.9SDD349 pKa = 3.5TFRR352 pKa = 11.84DD353 pKa = 4.26CVSLSAIASKK363 pKa = 10.25PFPLFHH369 pKa = 7.53IEE371 pKa = 3.88GFGYY375 pKa = 7.98YY376 pKa = 7.75QHH378 pKa = 6.49MRR380 pKa = 11.84FSDD383 pKa = 4.1LDD385 pKa = 3.63ATSATQLAITLDD397 pKa = 3.4YY398 pKa = 10.72HH399 pKa = 8.15LEE401 pKa = 4.04FRR403 pKa = 11.84SSSMLFNLGYY413 pKa = 10.55SSLPLEE419 pKa = 5.07AYY421 pKa = 10.39HH422 pKa = 6.37EE423 pKa = 4.43AQLALVKK430 pKa = 10.29MGTFFEE436 pKa = 4.47NPKK439 pKa = 10.19HH440 pKa = 6.38ISQITKK446 pKa = 10.06KK447 pKa = 10.56VLNSLPKK454 pKa = 9.41VRR456 pKa = 11.84APALVVMKK464 pKa = 8.43VTKK467 pKa = 10.47GAGRR471 pKa = 11.84PDD473 pKa = 3.51GSKK476 pKa = 10.73GGKK479 pKa = 8.94KK480 pKa = 10.3DD481 pKa = 3.65NAPKK485 pKa = 10.49QNGGKK490 pKa = 9.92GKK492 pKa = 10.18KK493 pKa = 9.13SKK495 pKa = 10.24KK496 pKa = 10.52GKK498 pKa = 9.75DD499 pKa = 3.06KK500 pKa = 11.03KK501 pKa = 10.95
Molecular weight: 54.19 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1648 |
430 |
717 |
549.3 |
60.37 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.83 ± 0.162 | 1.881 ± 0.419 |
4.187 ± 0.641 | 5.34 ± 0.689 |
3.883 ± 0.757 | 7.403 ± 0.697 |
1.881 ± 0.113 | 2.973 ± 0.602 |
4.551 ± 0.421 | 8.859 ± 0.362 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.699 ± 0.304 | 2.973 ± 0.448 |
7.767 ± 1.151 | 3.58 ± 0.386 |
5.583 ± 0.839 | 7.646 ± 0.547 |
6.25 ± 0.75 | 7.706 ± 0.403 |
2.549 ± 0.519 | 3.459 ± 0.567 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
