
Alteraurantiacibacter aquimixticola
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Sphingomonadales; Erythrobacteraceae; Alteraurantiacibacter
Average proteome isoelectric point is 5.74
Get precalculated fractions of proteins
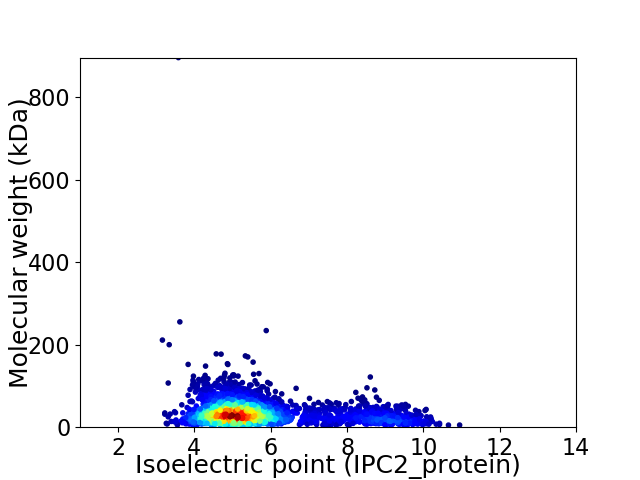
Virtual 2D-PAGE plot for 3188 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4T3EYD3|A0A4T3EYD3_9SPHN Type VI secretion protein OS=Alteraurantiacibacter aquimixticola OX=2489173 GN=E5222_12340 PE=4 SV=1
MM1 pKa = 7.74KK2 pKa = 10.33YY3 pKa = 10.86DD4 pKa = 3.62NLLAASLAAALVAACGGGSSGGGISSTPPPPAAPTPSPTSTSAPTPTPTNSNVGNLVADD63 pKa = 4.12EE64 pKa = 4.69AFSTLSSGSDD74 pKa = 2.79ISLSVIDD81 pKa = 4.79AEE83 pKa = 4.46VGGASVEE90 pKa = 4.05DD91 pKa = 3.53RR92 pKa = 11.84KK93 pKa = 10.89VAISYY98 pKa = 8.49DD99 pKa = 3.55ASTDD103 pKa = 3.33GYY105 pKa = 8.53TVSLGGQGSTFTSAGIIEE123 pKa = 4.21QGEE126 pKa = 4.48YY127 pKa = 10.29PGTTRR132 pKa = 11.84YY133 pKa = 8.24YY134 pKa = 9.21TEE136 pKa = 4.76SGNSSEE142 pKa = 4.88FLTISTRR149 pKa = 11.84GYY151 pKa = 9.31SQNSANQYY159 pKa = 8.83VALGYY164 pKa = 7.42WQRR167 pKa = 11.84NVLNGVQQEE176 pKa = 4.72TEE178 pKa = 3.26FDD180 pKa = 3.55TFVFGFPASSSAVPVSGLGSYY201 pKa = 9.91TIDD204 pKa = 2.82IFGALSVVGTEE215 pKa = 3.96ARR217 pKa = 11.84ALSGSGLLEE226 pKa = 4.31LDD228 pKa = 4.2FGTGAWRR235 pKa = 11.84VRR237 pKa = 11.84TTLGEE242 pKa = 4.2FPLADD247 pKa = 3.47YY248 pKa = 10.52GYY250 pKa = 11.15LSGGSLKK257 pKa = 10.61FNADD261 pKa = 3.08GFVTSNRR268 pKa = 11.84RR269 pKa = 11.84LDD271 pKa = 4.6GIFTYY276 pKa = 10.79TNLDD280 pKa = 3.24GDD282 pKa = 4.35FVSGDD287 pKa = 3.35VSGQFYY293 pKa = 10.81GPAAQEE299 pKa = 3.65VGAAFSGGTGTGAVLNGAFTGKK321 pKa = 9.89RR322 pKa = 11.84DD323 pKa = 3.61NQSRR327 pKa = 11.84AGTLALTNIQGDD339 pKa = 4.43TVVSGYY345 pKa = 10.27LAEE348 pKa = 4.53ALGTIQDD355 pKa = 4.29DD356 pKa = 3.65LTPAFRR362 pKa = 11.84GFYY365 pKa = 10.25GNTYY369 pKa = 7.21TEE371 pKa = 4.25RR372 pKa = 11.84PFTRR376 pKa = 11.84IHH378 pKa = 6.02FQQDD382 pKa = 2.97GTINVQLDD390 pKa = 3.7GALVASIEE398 pKa = 4.07PGEE401 pKa = 4.27LVSSDD406 pKa = 3.04GAYY409 pKa = 10.29DD410 pKa = 3.61VYY412 pKa = 10.92EE413 pKa = 4.31TTTTSAFGTAGLPLTARR430 pKa = 11.84IFKK433 pKa = 10.19PGTANPEE440 pKa = 3.89VQLTYY445 pKa = 11.1SSFGILEE452 pKa = 4.03QEE454 pKa = 4.22ISGSDD459 pKa = 3.43YY460 pKa = 9.64LQQVRR465 pKa = 11.84RR466 pKa = 11.84YY467 pKa = 6.94FTYY470 pKa = 10.54GFDD473 pKa = 3.59TPEE476 pKa = 3.22GHH478 pKa = 6.3MNVRR482 pKa = 11.84SGSASYY488 pKa = 10.86SGIAVGTTTNSSGLAIDD505 pKa = 3.73VDD507 pKa = 4.44GTSSFVVNFSSDD519 pKa = 3.5SYY521 pKa = 11.93SGMLDD526 pKa = 3.66LQADD530 pKa = 4.02DD531 pKa = 4.69GGTIVDD537 pKa = 4.44LGTFEE542 pKa = 4.39FASTITNGIMDD553 pKa = 5.57AANFDD558 pKa = 4.14YY559 pKa = 8.57PTALSASVQSLNTIIPVFYY578 pKa = 10.55GPDD581 pKa = 3.35GSEE584 pKa = 3.37IVAPFSILRR593 pKa = 11.84GPPNAIGTTTITGVAVAIEE612 pKa = 4.09DD613 pKa = 3.57
MM1 pKa = 7.74KK2 pKa = 10.33YY3 pKa = 10.86DD4 pKa = 3.62NLLAASLAAALVAACGGGSSGGGISSTPPPPAAPTPSPTSTSAPTPTPTNSNVGNLVADD63 pKa = 4.12EE64 pKa = 4.69AFSTLSSGSDD74 pKa = 2.79ISLSVIDD81 pKa = 4.79AEE83 pKa = 4.46VGGASVEE90 pKa = 4.05DD91 pKa = 3.53RR92 pKa = 11.84KK93 pKa = 10.89VAISYY98 pKa = 8.49DD99 pKa = 3.55ASTDD103 pKa = 3.33GYY105 pKa = 8.53TVSLGGQGSTFTSAGIIEE123 pKa = 4.21QGEE126 pKa = 4.48YY127 pKa = 10.29PGTTRR132 pKa = 11.84YY133 pKa = 8.24YY134 pKa = 9.21TEE136 pKa = 4.76SGNSSEE142 pKa = 4.88FLTISTRR149 pKa = 11.84GYY151 pKa = 9.31SQNSANQYY159 pKa = 8.83VALGYY164 pKa = 7.42WQRR167 pKa = 11.84NVLNGVQQEE176 pKa = 4.72TEE178 pKa = 3.26FDD180 pKa = 3.55TFVFGFPASSSAVPVSGLGSYY201 pKa = 9.91TIDD204 pKa = 2.82IFGALSVVGTEE215 pKa = 3.96ARR217 pKa = 11.84ALSGSGLLEE226 pKa = 4.31LDD228 pKa = 4.2FGTGAWRR235 pKa = 11.84VRR237 pKa = 11.84TTLGEE242 pKa = 4.2FPLADD247 pKa = 3.47YY248 pKa = 10.52GYY250 pKa = 11.15LSGGSLKK257 pKa = 10.61FNADD261 pKa = 3.08GFVTSNRR268 pKa = 11.84RR269 pKa = 11.84LDD271 pKa = 4.6GIFTYY276 pKa = 10.79TNLDD280 pKa = 3.24GDD282 pKa = 4.35FVSGDD287 pKa = 3.35VSGQFYY293 pKa = 10.81GPAAQEE299 pKa = 3.65VGAAFSGGTGTGAVLNGAFTGKK321 pKa = 9.89RR322 pKa = 11.84DD323 pKa = 3.61NQSRR327 pKa = 11.84AGTLALTNIQGDD339 pKa = 4.43TVVSGYY345 pKa = 10.27LAEE348 pKa = 4.53ALGTIQDD355 pKa = 4.29DD356 pKa = 3.65LTPAFRR362 pKa = 11.84GFYY365 pKa = 10.25GNTYY369 pKa = 7.21TEE371 pKa = 4.25RR372 pKa = 11.84PFTRR376 pKa = 11.84IHH378 pKa = 6.02FQQDD382 pKa = 2.97GTINVQLDD390 pKa = 3.7GALVASIEE398 pKa = 4.07PGEE401 pKa = 4.27LVSSDD406 pKa = 3.04GAYY409 pKa = 10.29DD410 pKa = 3.61VYY412 pKa = 10.92EE413 pKa = 4.31TTTTSAFGTAGLPLTARR430 pKa = 11.84IFKK433 pKa = 10.19PGTANPEE440 pKa = 3.89VQLTYY445 pKa = 11.1SSFGILEE452 pKa = 4.03QEE454 pKa = 4.22ISGSDD459 pKa = 3.43YY460 pKa = 9.64LQQVRR465 pKa = 11.84RR466 pKa = 11.84YY467 pKa = 6.94FTYY470 pKa = 10.54GFDD473 pKa = 3.59TPEE476 pKa = 3.22GHH478 pKa = 6.3MNVRR482 pKa = 11.84SGSASYY488 pKa = 10.86SGIAVGTTTNSSGLAIDD505 pKa = 3.73VDD507 pKa = 4.44GTSSFVVNFSSDD519 pKa = 3.5SYY521 pKa = 11.93SGMLDD526 pKa = 3.66LQADD530 pKa = 4.02DD531 pKa = 4.69GGTIVDD537 pKa = 4.44LGTFEE542 pKa = 4.39FASTITNGIMDD553 pKa = 5.57AANFDD558 pKa = 4.14YY559 pKa = 8.57PTALSASVQSLNTIIPVFYY578 pKa = 10.55GPDD581 pKa = 3.35GSEE584 pKa = 3.37IVAPFSILRR593 pKa = 11.84GPPNAIGTTTITGVAVAIEE612 pKa = 4.09DD613 pKa = 3.57
Molecular weight: 63.36 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4T3F646|A0A4T3F646_9SPHN Uncharacterized protein OS=Alteraurantiacibacter aquimixticola OX=2489173 GN=E5222_05440 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.89GFFARR21 pKa = 11.84KK22 pKa = 7.42ATPGGRR28 pKa = 11.84KK29 pKa = 7.93VLRR32 pKa = 11.84SRR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 8.51NLCAA44 pKa = 4.54
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.89GFFARR21 pKa = 11.84KK22 pKa = 7.42ATPGGRR28 pKa = 11.84KK29 pKa = 7.93VLRR32 pKa = 11.84SRR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 8.51NLCAA44 pKa = 4.54
Molecular weight: 5.2 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1045476 |
40 |
8505 |
327.9 |
35.57 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.388 ± 0.071 | 0.85 ± 0.015 |
6.247 ± 0.066 | 6.638 ± 0.051 |
3.689 ± 0.03 | 8.973 ± 0.053 |
1.951 ± 0.022 | 4.955 ± 0.028 |
2.854 ± 0.042 | 9.691 ± 0.058 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.596 ± 0.024 | 2.724 ± 0.032 |
5.137 ± 0.035 | 3.193 ± 0.024 |
6.746 ± 0.053 | 5.388 ± 0.032 |
5.159 ± 0.043 | 7.078 ± 0.029 |
1.468 ± 0.018 | 2.275 ± 0.026 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
