
Nitratireductor sp. SY7
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Phyllobacteriaceae; Nitratireductor; unclassified Nitratireductor
Average proteome isoelectric point is 6.34
Get precalculated fractions of proteins
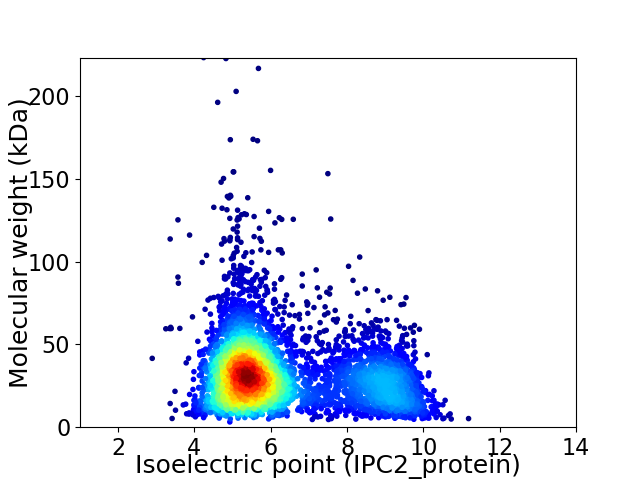
Virtual 2D-PAGE plot for 4636 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5B8KTH6|A0A5B8KTH6_9RHIZ Copper-bind domain-containing protein OS=Nitratireductor sp. SY7 OX=2599600 GN=FQ775_00110 PE=4 SV=1
MM1 pKa = 6.86QPAWNGSSEE10 pKa = 4.11PAGVFDD16 pKa = 5.32LTGDD20 pKa = 3.87PNIDD24 pKa = 3.76GILARR29 pKa = 11.84RR30 pKa = 11.84FWSDD34 pKa = 2.62GSITYY39 pKa = 9.32SFPDD43 pKa = 3.21QASDD47 pKa = 3.46YY48 pKa = 10.6GAYY51 pKa = 10.24SYY53 pKa = 11.95VDD55 pKa = 3.6GNGVTHH61 pKa = 7.08FPTATFQQVLAAQQTAIHH79 pKa = 6.43FAMSVEE85 pKa = 3.94NGPLASQGFSVEE97 pKa = 3.94GFTNLDD103 pKa = 3.25VGFNGTTTGNATMRR117 pKa = 11.84FGRR120 pKa = 11.84SDD122 pKa = 3.64DD123 pKa = 3.89AQPTAFAFYY132 pKa = 9.69PSSSNVGGDD141 pKa = 3.02VWLGTQVGGSNLSNPAAGNYY161 pKa = 8.54AWATILHH168 pKa = 6.68EE169 pKa = 4.88IGHH172 pKa = 6.13GLGLKK177 pKa = 9.73HH178 pKa = 6.0GHH180 pKa = 6.8AGDD183 pKa = 4.06SGFPNANPNVLPSNVDD199 pKa = 2.9AMEE202 pKa = 3.99YY203 pKa = 10.72SIMTYY208 pKa = 10.2RR209 pKa = 11.84SFINGPTNGYY219 pKa = 7.63TNEE222 pKa = 4.31TNGFAQTFMMLDD234 pKa = 3.61IAALQAMYY242 pKa = 10.39GADD245 pKa = 4.42FSSQSNNTDD254 pKa = 2.84TVYY257 pKa = 10.67KK258 pKa = 9.75WDD260 pKa = 3.83PTSGDD265 pKa = 3.54TLINGQVGIDD275 pKa = 3.28AAANRR280 pKa = 11.84IFATIWDD287 pKa = 4.04GGGVDD292 pKa = 3.97TYY294 pKa = 11.59DD295 pKa = 3.19LSSYY299 pKa = 9.54TGNLMLDD306 pKa = 3.9LRR308 pKa = 11.84PGEE311 pKa = 4.13EE312 pKa = 4.34SLFSTTQQAHH322 pKa = 6.38LGSGNFASGNIYY334 pKa = 10.53NALQFQGDD342 pKa = 3.8ARR344 pKa = 11.84SLIEE348 pKa = 3.9NAIGGTGNDD357 pKa = 4.11TITGNAANNSLFGGQGDD374 pKa = 4.05DD375 pKa = 3.8TLSGGNGNDD384 pKa = 3.76LLAGGAGVDD393 pKa = 3.78QINGEE398 pKa = 4.35GGVDD402 pKa = 3.26TADD405 pKa = 3.71YY406 pKa = 9.74SASAASVNVNLSTGTGSGGDD426 pKa = 3.65AQGDD430 pKa = 3.94TLTGIEE436 pKa = 4.4NLIGSNVAGTDD447 pKa = 3.31VLTGDD452 pKa = 3.78GTANHH457 pKa = 6.32IQGLAGNDD465 pKa = 3.09ILVGLGGADD474 pKa = 2.87ILVGGTGVDD483 pKa = 3.31TADD486 pKa = 3.59YY487 pKa = 9.23SASGGRR493 pKa = 11.84INVNLTTGAGLGGDD507 pKa = 3.44AHH509 pKa = 7.47GDD511 pKa = 3.54TLSGIEE517 pKa = 4.33NLIGTNIAMTDD528 pKa = 4.29FLTGDD533 pKa = 3.36AGANHH538 pKa = 6.37IQGLAGDD545 pKa = 4.65DD546 pKa = 4.56LIAGLGGADD555 pKa = 3.57VLIGGSGVDD564 pKa = 3.29TADD567 pKa = 3.54YY568 pKa = 10.12SASASRR574 pKa = 11.84IAVNLSTGYY583 pKa = 10.68AAGGDD588 pKa = 3.6AHH590 pKa = 7.44GDD592 pKa = 3.55TLSGIEE598 pKa = 4.3NLIGTNSSLTDD609 pKa = 3.97FLTGDD614 pKa = 3.48AGANHH619 pKa = 6.37IQGLAGDD626 pKa = 4.65DD627 pKa = 4.56LIAGLGGADD636 pKa = 3.57VLIGGSGVDD645 pKa = 3.29TADD648 pKa = 3.54YY649 pKa = 10.12SASASRR655 pKa = 11.84IAVNLSTGYY664 pKa = 10.68AAGGDD669 pKa = 3.6AHH671 pKa = 7.44GDD673 pKa = 3.55TLSGIEE679 pKa = 4.26NLIGTNVALTDD690 pKa = 4.06FLTGNSLANRR700 pKa = 11.84IEE702 pKa = 4.33GGAGDD707 pKa = 5.04DD708 pKa = 5.0LIAGLGGADD717 pKa = 3.57VLIGGSGVDD726 pKa = 3.29TADD729 pKa = 3.46YY730 pKa = 9.82SASGGRR736 pKa = 11.84IAVNLSTGYY745 pKa = 10.68AAGGDD750 pKa = 3.6AHH752 pKa = 7.44GDD754 pKa = 3.55TLSGIEE760 pKa = 4.26NLIGTNVALTDD771 pKa = 4.23FLTGDD776 pKa = 3.29AGANRR781 pKa = 11.84IEE783 pKa = 4.47GLAGDD788 pKa = 4.81DD789 pKa = 4.21EE790 pKa = 5.38INGKK794 pKa = 9.66EE795 pKa = 3.97GADD798 pKa = 3.13IFVGGGGDD806 pKa = 3.82DD807 pKa = 4.95LFIFDD812 pKa = 4.25TALGAGNIDD821 pKa = 4.53AILDD825 pKa = 3.67FVVADD830 pKa = 3.92DD831 pKa = 4.08MVRR834 pKa = 11.84LASSIFTGLSVGTLTAAAFRR854 pKa = 11.84VGAAAADD861 pKa = 3.66ADD863 pKa = 4.9DD864 pKa = 5.3RR865 pKa = 11.84IIYY868 pKa = 9.53NAGSGGLFFDD878 pKa = 3.86VDD880 pKa = 3.81GNGAQGQVQFASLSTGLALTNDD902 pKa = 3.84NFLVAA907 pKa = 5.3
MM1 pKa = 6.86QPAWNGSSEE10 pKa = 4.11PAGVFDD16 pKa = 5.32LTGDD20 pKa = 3.87PNIDD24 pKa = 3.76GILARR29 pKa = 11.84RR30 pKa = 11.84FWSDD34 pKa = 2.62GSITYY39 pKa = 9.32SFPDD43 pKa = 3.21QASDD47 pKa = 3.46YY48 pKa = 10.6GAYY51 pKa = 10.24SYY53 pKa = 11.95VDD55 pKa = 3.6GNGVTHH61 pKa = 7.08FPTATFQQVLAAQQTAIHH79 pKa = 6.43FAMSVEE85 pKa = 3.94NGPLASQGFSVEE97 pKa = 3.94GFTNLDD103 pKa = 3.25VGFNGTTTGNATMRR117 pKa = 11.84FGRR120 pKa = 11.84SDD122 pKa = 3.64DD123 pKa = 3.89AQPTAFAFYY132 pKa = 9.69PSSSNVGGDD141 pKa = 3.02VWLGTQVGGSNLSNPAAGNYY161 pKa = 8.54AWATILHH168 pKa = 6.68EE169 pKa = 4.88IGHH172 pKa = 6.13GLGLKK177 pKa = 9.73HH178 pKa = 6.0GHH180 pKa = 6.8AGDD183 pKa = 4.06SGFPNANPNVLPSNVDD199 pKa = 2.9AMEE202 pKa = 3.99YY203 pKa = 10.72SIMTYY208 pKa = 10.2RR209 pKa = 11.84SFINGPTNGYY219 pKa = 7.63TNEE222 pKa = 4.31TNGFAQTFMMLDD234 pKa = 3.61IAALQAMYY242 pKa = 10.39GADD245 pKa = 4.42FSSQSNNTDD254 pKa = 2.84TVYY257 pKa = 10.67KK258 pKa = 9.75WDD260 pKa = 3.83PTSGDD265 pKa = 3.54TLINGQVGIDD275 pKa = 3.28AAANRR280 pKa = 11.84IFATIWDD287 pKa = 4.04GGGVDD292 pKa = 3.97TYY294 pKa = 11.59DD295 pKa = 3.19LSSYY299 pKa = 9.54TGNLMLDD306 pKa = 3.9LRR308 pKa = 11.84PGEE311 pKa = 4.13EE312 pKa = 4.34SLFSTTQQAHH322 pKa = 6.38LGSGNFASGNIYY334 pKa = 10.53NALQFQGDD342 pKa = 3.8ARR344 pKa = 11.84SLIEE348 pKa = 3.9NAIGGTGNDD357 pKa = 4.11TITGNAANNSLFGGQGDD374 pKa = 4.05DD375 pKa = 3.8TLSGGNGNDD384 pKa = 3.76LLAGGAGVDD393 pKa = 3.78QINGEE398 pKa = 4.35GGVDD402 pKa = 3.26TADD405 pKa = 3.71YY406 pKa = 9.74SASAASVNVNLSTGTGSGGDD426 pKa = 3.65AQGDD430 pKa = 3.94TLTGIEE436 pKa = 4.4NLIGSNVAGTDD447 pKa = 3.31VLTGDD452 pKa = 3.78GTANHH457 pKa = 6.32IQGLAGNDD465 pKa = 3.09ILVGLGGADD474 pKa = 2.87ILVGGTGVDD483 pKa = 3.31TADD486 pKa = 3.59YY487 pKa = 9.23SASGGRR493 pKa = 11.84INVNLTTGAGLGGDD507 pKa = 3.44AHH509 pKa = 7.47GDD511 pKa = 3.54TLSGIEE517 pKa = 4.33NLIGTNIAMTDD528 pKa = 4.29FLTGDD533 pKa = 3.36AGANHH538 pKa = 6.37IQGLAGDD545 pKa = 4.65DD546 pKa = 4.56LIAGLGGADD555 pKa = 3.57VLIGGSGVDD564 pKa = 3.29TADD567 pKa = 3.54YY568 pKa = 10.12SASASRR574 pKa = 11.84IAVNLSTGYY583 pKa = 10.68AAGGDD588 pKa = 3.6AHH590 pKa = 7.44GDD592 pKa = 3.55TLSGIEE598 pKa = 4.3NLIGTNSSLTDD609 pKa = 3.97FLTGDD614 pKa = 3.48AGANHH619 pKa = 6.37IQGLAGDD626 pKa = 4.65DD627 pKa = 4.56LIAGLGGADD636 pKa = 3.57VLIGGSGVDD645 pKa = 3.29TADD648 pKa = 3.54YY649 pKa = 10.12SASASRR655 pKa = 11.84IAVNLSTGYY664 pKa = 10.68AAGGDD669 pKa = 3.6AHH671 pKa = 7.44GDD673 pKa = 3.55TLSGIEE679 pKa = 4.26NLIGTNVALTDD690 pKa = 4.06FLTGNSLANRR700 pKa = 11.84IEE702 pKa = 4.33GGAGDD707 pKa = 5.04DD708 pKa = 5.0LIAGLGGADD717 pKa = 3.57VLIGGSGVDD726 pKa = 3.29TADD729 pKa = 3.46YY730 pKa = 9.82SASGGRR736 pKa = 11.84IAVNLSTGYY745 pKa = 10.68AAGGDD750 pKa = 3.6AHH752 pKa = 7.44GDD754 pKa = 3.55TLSGIEE760 pKa = 4.26NLIGTNVALTDD771 pKa = 4.23FLTGDD776 pKa = 3.29AGANRR781 pKa = 11.84IEE783 pKa = 4.47GLAGDD788 pKa = 4.81DD789 pKa = 4.21EE790 pKa = 5.38INGKK794 pKa = 9.66EE795 pKa = 3.97GADD798 pKa = 3.13IFVGGGGDD806 pKa = 3.82DD807 pKa = 4.95LFIFDD812 pKa = 4.25TALGAGNIDD821 pKa = 4.53AILDD825 pKa = 3.67FVVADD830 pKa = 3.92DD831 pKa = 4.08MVRR834 pKa = 11.84LASSIFTGLSVGTLTAAAFRR854 pKa = 11.84VGAAAADD861 pKa = 3.66ADD863 pKa = 4.9DD864 pKa = 5.3RR865 pKa = 11.84IIYY868 pKa = 9.53NAGSGGLFFDD878 pKa = 3.86VDD880 pKa = 3.81GNGAQGQVQFASLSTGLALTNDD902 pKa = 3.84NFLVAA907 pKa = 5.3
Molecular weight: 90.6 kDa
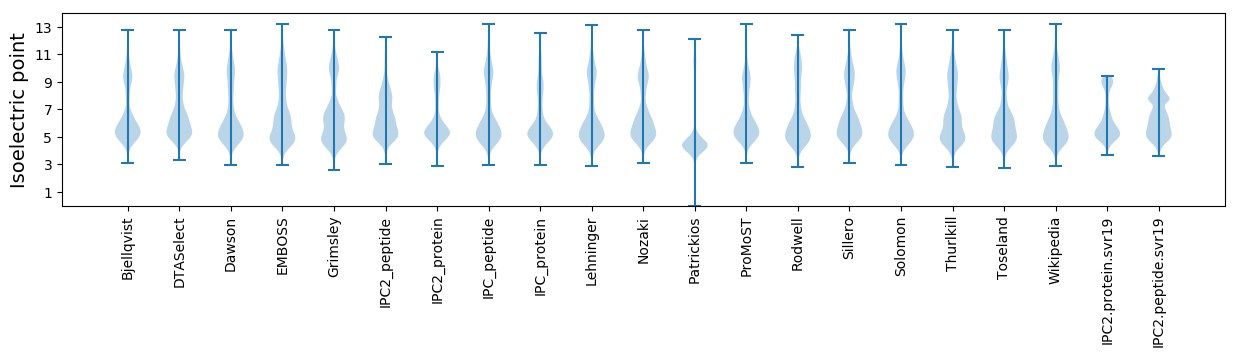
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5B8L058|A0A5B8L058_9RHIZ (S)-ureidoglycine aminohydrolase OS=Nitratireductor sp. SY7 OX=2599600 GN=FQ775_14065 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.73LVRR12 pKa = 11.84KK13 pKa = 9.15RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 9.68GGRR28 pKa = 11.84RR29 pKa = 11.84VVAARR34 pKa = 11.84RR35 pKa = 11.84NRR37 pKa = 11.84GRR39 pKa = 11.84NRR41 pKa = 11.84LSAA44 pKa = 3.96
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.73LVRR12 pKa = 11.84KK13 pKa = 9.15RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 9.68GGRR28 pKa = 11.84RR29 pKa = 11.84VVAARR34 pKa = 11.84RR35 pKa = 11.84NRR37 pKa = 11.84GRR39 pKa = 11.84NRR41 pKa = 11.84LSAA44 pKa = 3.96
Molecular weight: 5.21 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1446749 |
32 |
2124 |
312.1 |
33.83 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.926 ± 0.052 | 0.813 ± 0.011 |
5.844 ± 0.035 | 6.186 ± 0.037 |
3.925 ± 0.026 | 8.804 ± 0.034 |
2.018 ± 0.016 | 5.226 ± 0.026 |
3.097 ± 0.032 | 9.825 ± 0.044 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.518 ± 0.017 | 2.491 ± 0.022 |
4.983 ± 0.028 | 2.704 ± 0.018 |
7.376 ± 0.037 | 5.199 ± 0.021 |
5.031 ± 0.022 | 7.558 ± 0.028 |
1.311 ± 0.016 | 2.168 ± 0.016 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
