
Halalkalicoccus jeotgali (strain DSM 18796 / CECT 7217 / JCM 14584 / KCTC 4019 / B3)
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Stenosarchaea group; Halobacteria; Halobacteriales; Halobacteriaceae; Halalkalicoccus; Halalkalicoccus jeotgali
Average proteome isoelectric point is 5.02
Get precalculated fractions of proteins
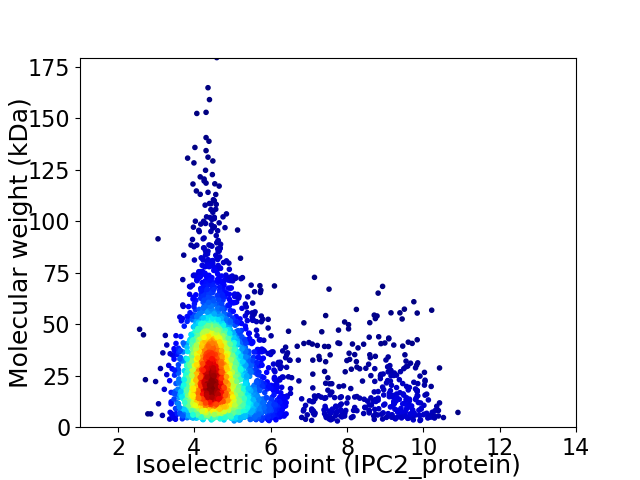
Virtual 2D-PAGE plot for 3779 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|D8JB89|D8JB89_HALJB ATP/cobalamin adenosyltransferase OS=Halalkalicoccus jeotgali (strain DSM 18796 / CECT 7217 / JCM 14584 / KCTC 4019 / B3) OX=795797 GN=HacjB3_15921 PE=4 SV=1
MM1 pKa = 7.88SGEE4 pKa = 3.81EE5 pKa = 3.81ALYY8 pKa = 10.75VVDD11 pKa = 5.94DD12 pKa = 4.07SGHH15 pKa = 4.79VLRR18 pKa = 11.84GVNNGGDD25 pKa = 3.95VTWDD29 pKa = 3.48EE30 pKa = 4.42PQTPGDD36 pKa = 3.84GSDD39 pKa = 2.94ITSVVFTDD47 pKa = 3.78EE48 pKa = 5.61AGFLCDD54 pKa = 3.54TNAGVYY60 pKa = 7.56EE61 pKa = 4.26TTDD64 pKa = 5.58DD65 pKa = 3.63GDD67 pKa = 3.23TWDD70 pKa = 4.54RR71 pKa = 11.84IGIEE75 pKa = 3.81EE76 pKa = 4.91AGPDD80 pKa = 3.66FTDD83 pKa = 3.37IAPARR88 pKa = 11.84RR89 pKa = 11.84DD90 pKa = 4.02AISVSCDD97 pKa = 2.99DD98 pKa = 3.86GTVFRR103 pKa = 11.84YY104 pKa = 10.46DD105 pKa = 3.06GTNWTNRR112 pKa = 11.84YY113 pKa = 9.06IGEE116 pKa = 4.14EE117 pKa = 3.99ALCAIDD123 pKa = 5.31RR124 pKa = 11.84NTDD127 pKa = 2.98TGLLCGEE134 pKa = 5.16DD135 pKa = 3.42GAIYY139 pKa = 9.15EE140 pKa = 4.25QQQGRR145 pKa = 11.84WEE147 pKa = 4.1RR148 pKa = 11.84QPTPIEE154 pKa = 4.11EE155 pKa = 4.32TLHH158 pKa = 6.39GIAIGTEE165 pKa = 4.19YY166 pKa = 10.43PSVAVGEE173 pKa = 4.65DD174 pKa = 3.41GTIIEE179 pKa = 4.65YY180 pKa = 10.36VV181 pKa = 3.13
MM1 pKa = 7.88SGEE4 pKa = 3.81EE5 pKa = 3.81ALYY8 pKa = 10.75VVDD11 pKa = 5.94DD12 pKa = 4.07SGHH15 pKa = 4.79VLRR18 pKa = 11.84GVNNGGDD25 pKa = 3.95VTWDD29 pKa = 3.48EE30 pKa = 4.42PQTPGDD36 pKa = 3.84GSDD39 pKa = 2.94ITSVVFTDD47 pKa = 3.78EE48 pKa = 5.61AGFLCDD54 pKa = 3.54TNAGVYY60 pKa = 7.56EE61 pKa = 4.26TTDD64 pKa = 5.58DD65 pKa = 3.63GDD67 pKa = 3.23TWDD70 pKa = 4.54RR71 pKa = 11.84IGIEE75 pKa = 3.81EE76 pKa = 4.91AGPDD80 pKa = 3.66FTDD83 pKa = 3.37IAPARR88 pKa = 11.84RR89 pKa = 11.84DD90 pKa = 4.02AISVSCDD97 pKa = 2.99DD98 pKa = 3.86GTVFRR103 pKa = 11.84YY104 pKa = 10.46DD105 pKa = 3.06GTNWTNRR112 pKa = 11.84YY113 pKa = 9.06IGEE116 pKa = 4.14EE117 pKa = 3.99ALCAIDD123 pKa = 5.31RR124 pKa = 11.84NTDD127 pKa = 2.98TGLLCGEE134 pKa = 5.16DD135 pKa = 3.42GAIYY139 pKa = 9.15EE140 pKa = 4.25QQQGRR145 pKa = 11.84WEE147 pKa = 4.1RR148 pKa = 11.84QPTPIEE154 pKa = 4.11EE155 pKa = 4.32TLHH158 pKa = 6.39GIAIGTEE165 pKa = 4.19YY166 pKa = 10.43PSVAVGEE173 pKa = 4.65DD174 pKa = 3.41GTIIEE179 pKa = 4.65YY180 pKa = 10.36VV181 pKa = 3.13
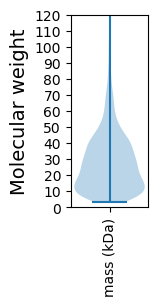
Molecular weight: 19.6 kDa
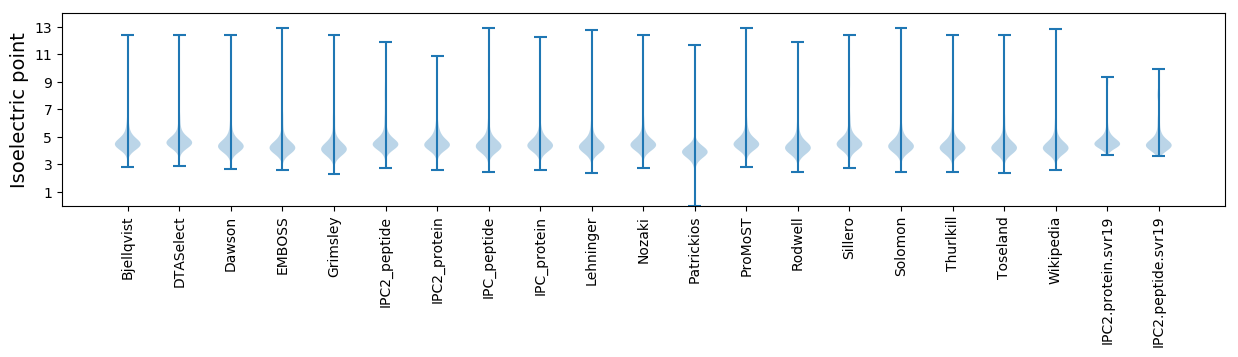
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|D8J297|D8J297_HALJB Sodium/hydrogen exchanger OS=Halalkalicoccus jeotgali (strain DSM 18796 / CECT 7217 / JCM 14584 / KCTC 4019 / B3) OX=795797 GN=HacjB3_07345 PE=4 SV=1
MM1 pKa = 7.81ILDD4 pKa = 4.01SLLQNFMMRR13 pKa = 11.84SCLMRR18 pKa = 11.84NSAAPISIEE27 pKa = 3.99RR28 pKa = 11.84SSKK31 pKa = 10.95ALISLVRR38 pKa = 11.84TLLSTLLVGLTILAQSSMILTYY60 pKa = 9.55FMRR63 pKa = 11.84IFVVLNSLTWCFATRR78 pKa = 11.84ISLEE82 pKa = 4.22VIILTLTLEE91 pKa = 3.98MLILVMQHH99 pKa = 6.08FIMRR103 pKa = 11.84FSHH106 pKa = 6.33ALIVAEE112 pKa = 4.2RR113 pKa = 11.84HH114 pKa = 5.71LPMLYY119 pKa = 10.38FMRR122 pKa = 11.84RR123 pKa = 11.84FLQMLGLTRR132 pKa = 11.84KK133 pKa = 8.31RR134 pKa = 11.84HH135 pKa = 4.71SMIQAFLDD143 pKa = 3.1QWLSMKK149 pKa = 8.66KK150 pKa = 7.09THH152 pKa = 6.27MLWKK156 pKa = 9.46DD157 pKa = 3.76CPWILIHH164 pKa = 7.31
MM1 pKa = 7.81ILDD4 pKa = 4.01SLLQNFMMRR13 pKa = 11.84SCLMRR18 pKa = 11.84NSAAPISIEE27 pKa = 3.99RR28 pKa = 11.84SSKK31 pKa = 10.95ALISLVRR38 pKa = 11.84TLLSTLLVGLTILAQSSMILTYY60 pKa = 9.55FMRR63 pKa = 11.84IFVVLNSLTWCFATRR78 pKa = 11.84ISLEE82 pKa = 4.22VIILTLTLEE91 pKa = 3.98MLILVMQHH99 pKa = 6.08FIMRR103 pKa = 11.84FSHH106 pKa = 6.33ALIVAEE112 pKa = 4.2RR113 pKa = 11.84HH114 pKa = 5.71LPMLYY119 pKa = 10.38FMRR122 pKa = 11.84RR123 pKa = 11.84FLQMLGLTRR132 pKa = 11.84KK133 pKa = 8.31RR134 pKa = 11.84HH135 pKa = 4.71SMIQAFLDD143 pKa = 3.1QWLSMKK149 pKa = 8.66KK150 pKa = 7.09THH152 pKa = 6.27MLWKK156 pKa = 9.46DD157 pKa = 3.76CPWILIHH164 pKa = 7.31
Molecular weight: 19.31 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1011784 |
30 |
1575 |
267.7 |
29.2 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.477 ± 0.046 | 0.772 ± 0.013 |
7.393 ± 0.05 | 9.176 ± 0.064 |
3.42 ± 0.028 | 8.784 ± 0.048 |
2.032 ± 0.024 | 4.68 ± 0.033 |
1.882 ± 0.024 | 9.177 ± 0.051 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.898 ± 0.017 | 2.361 ± 0.024 |
4.691 ± 0.027 | 2.362 ± 0.025 |
6.688 ± 0.048 | 5.517 ± 0.029 |
6.231 ± 0.031 | 8.497 ± 0.045 |
1.152 ± 0.015 | 2.81 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
