
Fomitopsis rosea
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Basidiomycota; Agaricomycotina; Agaricomycetes; Agaricomycetes incertae sedis; Polyporales; Fomitopsidaceae; Fomitopsis
Average proteome isoelectric point is 6.37
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 11196 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
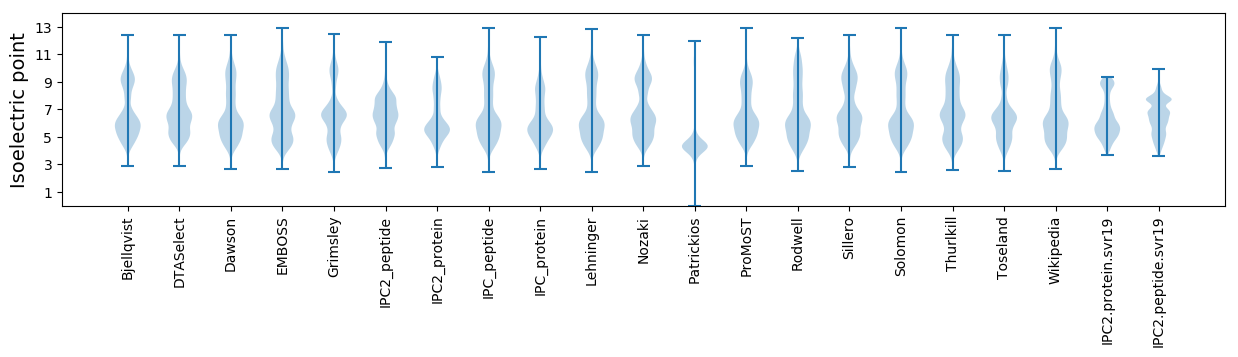
* You can choose from 21 different methods for calculating isoelectric point
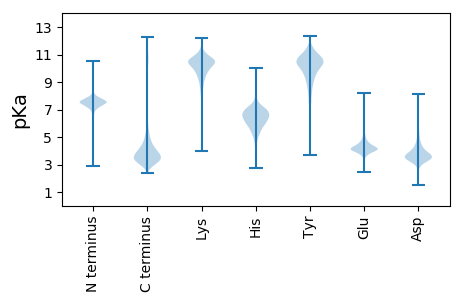
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A4Y9YJ24|A0A4Y9YJ24_9APHY Peptidase S53 domain-containing protein OS=Fomitopsis rosea OX=34475 GN=EVJ58_g4155 PE=4 SV=1
MM1 pKa = 6.85NTFFFGLLLASLCGLTNALYY21 pKa = 9.64IPVPVRR27 pKa = 11.84HH28 pKa = 6.7DD29 pKa = 3.48AQVSSRR35 pKa = 11.84GTGVHH40 pKa = 5.92NPAIRR45 pKa = 11.84LVGAFRR51 pKa = 11.84QQDD54 pKa = 3.51ANNDD58 pKa = 3.75SVVNFQDD65 pKa = 3.14TVYY68 pKa = 9.36ATNMTIGGQDD78 pKa = 4.28LLIQLDD84 pKa = 4.04TGSSDD89 pKa = 2.75LWVNLEE95 pKa = 3.9AGEE98 pKa = 4.55IEE100 pKa = 4.52FTNTTDD106 pKa = 3.86LPAEE110 pKa = 3.8EE111 pKa = 5.32AYY113 pKa = 10.56GIGHH117 pKa = 6.23ITGTIQFANAEE128 pKa = 4.27LGDD131 pKa = 4.23NIVPNQAFMNVSNATSFSTIFNDD154 pKa = 4.17GVRR157 pKa = 11.84GILGLAFDD165 pKa = 5.15AGSTVQGTIAQVYY178 pKa = 10.22GEE180 pKa = 4.97DD181 pKa = 3.65STLGRR186 pKa = 11.84SFLSNLFNQNTTKK199 pKa = 10.68PLFTVQLGRR208 pKa = 11.84TDD210 pKa = 3.84DD211 pKa = 3.88PQYY214 pKa = 9.14TAEE217 pKa = 4.11GAFTIGEE224 pKa = 4.28YY225 pKa = 10.82LPDD228 pKa = 3.83YY229 pKa = 11.32DD230 pKa = 5.2EE231 pKa = 4.73VQDD234 pKa = 3.72MPKK237 pKa = 10.41LHH239 pKa = 7.1RR240 pKa = 11.84FPNNGTGAAPRR251 pKa = 11.84WSTVMSGMTINGEE264 pKa = 3.84PFQFNASGVPDD275 pKa = 3.94TPEE278 pKa = 4.28GSVVAVLDD286 pKa = 3.59TGFTFPPIPGPAVEE300 pKa = 5.51AIYY303 pKa = 10.8SGIQGAYY310 pKa = 9.45YY311 pKa = 8.51DD312 pKa = 4.57TNSSLWIVPCNKK324 pKa = 9.6AANVEE329 pKa = 4.35FQFGDD334 pKa = 3.34MSVPIHH340 pKa = 6.54PLDD343 pKa = 3.42LTTIVTINNTEE354 pKa = 3.85VCVNTFRR361 pKa = 11.84PSTFPVNNQFDD372 pKa = 4.7LVLGDD377 pKa = 4.55AFLKK381 pKa = 10.3NVYY384 pKa = 10.13VSFNYY389 pKa = 10.86GDD391 pKa = 3.7MSADD395 pKa = 3.16AVPFMQIMPTTDD407 pKa = 2.39MDD409 pKa = 4.18AATKK413 pKa = 10.23EE414 pKa = 4.14FASVRR419 pKa = 11.84GAMYY423 pKa = 10.97AEE425 pKa = 4.6AAASAAASSASASAASAYY443 pKa = 10.08AALEE447 pKa = 4.36SGSASVPCNAGAPTTTPLFTSAWSSSPSAVMTDD480 pKa = 3.4SAPANTDD487 pKa = 3.2SPSLYY492 pKa = 10.46DD493 pKa = 3.83PSSGDD498 pKa = 3.34GSSDD502 pKa = 3.18ATPSSSADD510 pKa = 3.09ASSATDD516 pKa = 3.26ASSPDD521 pKa = 3.65SASPAATTDD530 pKa = 3.11AATDD534 pKa = 3.58ASSTDD539 pKa = 3.24SSSPTTTSDD548 pKa = 3.72ASSTDD553 pKa = 3.19ASATATDD560 pKa = 3.96ASADD564 pKa = 3.58ATATPTDD571 pKa = 3.75ASSPTATSDD580 pKa = 3.67ASDD583 pKa = 3.61ASSTDD588 pKa = 3.2SSASTTDD595 pKa = 2.82ASTDD599 pKa = 3.24ATATTTDD606 pKa = 3.07ASADD610 pKa = 3.52ATATPTDD617 pKa = 3.87TPADD621 pKa = 3.53AATTTDD627 pKa = 3.42ASATSTDD634 pKa = 3.14SSADD638 pKa = 3.29ATATPDD644 pKa = 3.7DD645 pKa = 4.54SSDD648 pKa = 3.57NSSTGSLPIDD658 pKa = 4.2PSASADD664 pKa = 3.76DD665 pKa = 4.61ASSSSDD671 pKa = 3.57DD672 pKa = 3.7GSGSLSEE679 pKa = 4.21NLSLLGSPEE688 pKa = 4.19SKK690 pKa = 10.21QSSFGGNLYY699 pKa = 10.64GSNDD703 pKa = 3.34YY704 pKa = 10.68EE705 pKa = 4.07YY706 pKa = 10.9SRR708 pKa = 11.84RR709 pKa = 11.84FRR711 pKa = 11.84RR712 pKa = 11.84SYY714 pKa = 10.88RR715 pKa = 11.84EE716 pKa = 3.58PTKK719 pKa = 10.27TVHH722 pKa = 6.21EE723 pKa = 4.45CLAPLIEE730 pKa = 5.66DD731 pKa = 3.3YY732 pKa = 11.42GAFLFALIAGTFIISFALCVVTVFLAIRR760 pKa = 11.84NHH762 pKa = 4.6TRR764 pKa = 11.84RR765 pKa = 11.84DD766 pKa = 3.19ARR768 pKa = 11.84KK769 pKa = 7.95RR770 pKa = 11.84AAYY773 pKa = 9.8LVLEE777 pKa = 4.66ANAEE781 pKa = 4.07AEE783 pKa = 4.38ADD785 pKa = 3.96PEE787 pKa = 4.13AAAYY791 pKa = 7.1TATYY795 pKa = 9.86HH796 pKa = 5.37YY797 pKa = 9.57TDD799 pKa = 5.13DD800 pKa = 3.95YY801 pKa = 11.95
MM1 pKa = 6.85NTFFFGLLLASLCGLTNALYY21 pKa = 9.64IPVPVRR27 pKa = 11.84HH28 pKa = 6.7DD29 pKa = 3.48AQVSSRR35 pKa = 11.84GTGVHH40 pKa = 5.92NPAIRR45 pKa = 11.84LVGAFRR51 pKa = 11.84QQDD54 pKa = 3.51ANNDD58 pKa = 3.75SVVNFQDD65 pKa = 3.14TVYY68 pKa = 9.36ATNMTIGGQDD78 pKa = 4.28LLIQLDD84 pKa = 4.04TGSSDD89 pKa = 2.75LWVNLEE95 pKa = 3.9AGEE98 pKa = 4.55IEE100 pKa = 4.52FTNTTDD106 pKa = 3.86LPAEE110 pKa = 3.8EE111 pKa = 5.32AYY113 pKa = 10.56GIGHH117 pKa = 6.23ITGTIQFANAEE128 pKa = 4.27LGDD131 pKa = 4.23NIVPNQAFMNVSNATSFSTIFNDD154 pKa = 4.17GVRR157 pKa = 11.84GILGLAFDD165 pKa = 5.15AGSTVQGTIAQVYY178 pKa = 10.22GEE180 pKa = 4.97DD181 pKa = 3.65STLGRR186 pKa = 11.84SFLSNLFNQNTTKK199 pKa = 10.68PLFTVQLGRR208 pKa = 11.84TDD210 pKa = 3.84DD211 pKa = 3.88PQYY214 pKa = 9.14TAEE217 pKa = 4.11GAFTIGEE224 pKa = 4.28YY225 pKa = 10.82LPDD228 pKa = 3.83YY229 pKa = 11.32DD230 pKa = 5.2EE231 pKa = 4.73VQDD234 pKa = 3.72MPKK237 pKa = 10.41LHH239 pKa = 7.1RR240 pKa = 11.84FPNNGTGAAPRR251 pKa = 11.84WSTVMSGMTINGEE264 pKa = 3.84PFQFNASGVPDD275 pKa = 3.94TPEE278 pKa = 4.28GSVVAVLDD286 pKa = 3.59TGFTFPPIPGPAVEE300 pKa = 5.51AIYY303 pKa = 10.8SGIQGAYY310 pKa = 9.45YY311 pKa = 8.51DD312 pKa = 4.57TNSSLWIVPCNKK324 pKa = 9.6AANVEE329 pKa = 4.35FQFGDD334 pKa = 3.34MSVPIHH340 pKa = 6.54PLDD343 pKa = 3.42LTTIVTINNTEE354 pKa = 3.85VCVNTFRR361 pKa = 11.84PSTFPVNNQFDD372 pKa = 4.7LVLGDD377 pKa = 4.55AFLKK381 pKa = 10.3NVYY384 pKa = 10.13VSFNYY389 pKa = 10.86GDD391 pKa = 3.7MSADD395 pKa = 3.16AVPFMQIMPTTDD407 pKa = 2.39MDD409 pKa = 4.18AATKK413 pKa = 10.23EE414 pKa = 4.14FASVRR419 pKa = 11.84GAMYY423 pKa = 10.97AEE425 pKa = 4.6AAASAAASSASASAASAYY443 pKa = 10.08AALEE447 pKa = 4.36SGSASVPCNAGAPTTTPLFTSAWSSSPSAVMTDD480 pKa = 3.4SAPANTDD487 pKa = 3.2SPSLYY492 pKa = 10.46DD493 pKa = 3.83PSSGDD498 pKa = 3.34GSSDD502 pKa = 3.18ATPSSSADD510 pKa = 3.09ASSATDD516 pKa = 3.26ASSPDD521 pKa = 3.65SASPAATTDD530 pKa = 3.11AATDD534 pKa = 3.58ASSTDD539 pKa = 3.24SSSPTTTSDD548 pKa = 3.72ASSTDD553 pKa = 3.19ASATATDD560 pKa = 3.96ASADD564 pKa = 3.58ATATPTDD571 pKa = 3.75ASSPTATSDD580 pKa = 3.67ASDD583 pKa = 3.61ASSTDD588 pKa = 3.2SSASTTDD595 pKa = 2.82ASTDD599 pKa = 3.24ATATTTDD606 pKa = 3.07ASADD610 pKa = 3.52ATATPTDD617 pKa = 3.87TPADD621 pKa = 3.53AATTTDD627 pKa = 3.42ASATSTDD634 pKa = 3.14SSADD638 pKa = 3.29ATATPDD644 pKa = 3.7DD645 pKa = 4.54SSDD648 pKa = 3.57NSSTGSLPIDD658 pKa = 4.2PSASADD664 pKa = 3.76DD665 pKa = 4.61ASSSSDD671 pKa = 3.57DD672 pKa = 3.7GSGSLSEE679 pKa = 4.21NLSLLGSPEE688 pKa = 4.19SKK690 pKa = 10.21QSSFGGNLYY699 pKa = 10.64GSNDD703 pKa = 3.34YY704 pKa = 10.68EE705 pKa = 4.07YY706 pKa = 10.9SRR708 pKa = 11.84RR709 pKa = 11.84FRR711 pKa = 11.84RR712 pKa = 11.84SYY714 pKa = 10.88RR715 pKa = 11.84EE716 pKa = 3.58PTKK719 pKa = 10.27TVHH722 pKa = 6.21EE723 pKa = 4.45CLAPLIEE730 pKa = 5.66DD731 pKa = 3.3YY732 pKa = 11.42GAFLFALIAGTFIISFALCVVTVFLAIRR760 pKa = 11.84NHH762 pKa = 4.6TRR764 pKa = 11.84RR765 pKa = 11.84DD766 pKa = 3.19ARR768 pKa = 11.84KK769 pKa = 7.95RR770 pKa = 11.84AAYY773 pKa = 9.8LVLEE777 pKa = 4.66ANAEE781 pKa = 4.07AEE783 pKa = 4.38ADD785 pKa = 3.96PEE787 pKa = 4.13AAAYY791 pKa = 7.1TATYY795 pKa = 9.86HH796 pKa = 5.37YY797 pKa = 9.57TDD799 pKa = 5.13DD800 pKa = 3.95YY801 pKa = 11.95
Molecular weight: 83.02 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4Y9XX56|A0A4Y9XX56_9APHY Uncharacterized protein OS=Fomitopsis rosea OX=34475 GN=EVJ58_g8964 PE=4 SV=1
MM1 pKa = 7.3TLLTKK6 pKa = 10.87SMAPGADD13 pKa = 3.48SQIGTSARR21 pKa = 11.84RR22 pKa = 11.84HH23 pKa = 4.41RR24 pKa = 11.84TAVGAPIVRR33 pKa = 11.84NCAHH37 pKa = 6.73RR38 pKa = 11.84RR39 pKa = 11.84HH40 pKa = 6.69PKK42 pKa = 8.34RR43 pKa = 11.84THH45 pKa = 6.36ASTTRR50 pKa = 11.84RR51 pKa = 11.84RR52 pKa = 11.84RR53 pKa = 11.84RR54 pKa = 11.84PDD56 pKa = 2.73ARR58 pKa = 11.84RR59 pKa = 11.84GWSSRR64 pKa = 11.84CRR66 pKa = 11.84ASASLRR72 pKa = 11.84RR73 pKa = 11.84VRR75 pKa = 11.84WSSSRR80 pKa = 11.84RR81 pKa = 11.84RR82 pKa = 11.84STGGRR87 pKa = 11.84SRR89 pKa = 11.84ASGTRR94 pKa = 11.84SASPARR100 pKa = 11.84RR101 pKa = 11.84GKK103 pKa = 10.32SSPFACSTLNPYY115 pKa = 8.2ITILLTFLATAXKK128 pKa = 9.74DD129 pKa = 3.48AQALXVLEE137 pKa = 5.4RR138 pKa = 11.84FIPWADD144 pKa = 3.24LAVFLTAIPRR154 pKa = 11.84RR155 pKa = 11.84VTYY158 pKa = 10.49RR159 pKa = 11.84EE160 pKa = 3.9QQKK163 pKa = 9.77EE164 pKa = 4.06RR165 pKa = 11.84SDD167 pKa = 3.74SAVLLTSGCKK177 pKa = 9.46PLPEE181 pKa = 4.38DD182 pKa = 2.88WCLRR186 pKa = 11.84GLGWGGKK193 pKa = 8.16RR194 pKa = 11.84VYY196 pKa = 10.97PMGFWGKK203 pKa = 8.76EE204 pKa = 3.67ANVEE208 pKa = 3.98DD209 pKa = 4.64RR210 pKa = 11.84NVEE213 pKa = 4.03VEE215 pKa = 4.3VLDD218 pKa = 3.96KK219 pKa = 11.68SEE221 pKa = 5.97GGDD224 pKa = 3.3QLDD227 pKa = 5.14GIIEE231 pKa = 4.11DD232 pKa = 3.46EE233 pKa = 4.38RR234 pKa = 11.84EE235 pKa = 3.86EE236 pKa = 4.17NEE238 pKa = 4.24AANEE242 pKa = 4.42LGKK245 pKa = 10.69CWIRR249 pKa = 11.84VACAGLKK256 pKa = 8.95IAKK259 pKa = 9.39HH260 pKa = 5.42VGGFAYY266 pKa = 10.5YY267 pKa = 9.05PAANEE272 pKa = 3.91EE273 pKa = 4.28HH274 pKa = 7.16RR275 pKa = 11.84GQQ277 pKa = 3.8
MM1 pKa = 7.3TLLTKK6 pKa = 10.87SMAPGADD13 pKa = 3.48SQIGTSARR21 pKa = 11.84RR22 pKa = 11.84HH23 pKa = 4.41RR24 pKa = 11.84TAVGAPIVRR33 pKa = 11.84NCAHH37 pKa = 6.73RR38 pKa = 11.84RR39 pKa = 11.84HH40 pKa = 6.69PKK42 pKa = 8.34RR43 pKa = 11.84THH45 pKa = 6.36ASTTRR50 pKa = 11.84RR51 pKa = 11.84RR52 pKa = 11.84RR53 pKa = 11.84RR54 pKa = 11.84PDD56 pKa = 2.73ARR58 pKa = 11.84RR59 pKa = 11.84GWSSRR64 pKa = 11.84CRR66 pKa = 11.84ASASLRR72 pKa = 11.84RR73 pKa = 11.84VRR75 pKa = 11.84WSSSRR80 pKa = 11.84RR81 pKa = 11.84RR82 pKa = 11.84STGGRR87 pKa = 11.84SRR89 pKa = 11.84ASGTRR94 pKa = 11.84SASPARR100 pKa = 11.84RR101 pKa = 11.84GKK103 pKa = 10.32SSPFACSTLNPYY115 pKa = 8.2ITILLTFLATAXKK128 pKa = 9.74DD129 pKa = 3.48AQALXVLEE137 pKa = 5.4RR138 pKa = 11.84FIPWADD144 pKa = 3.24LAVFLTAIPRR154 pKa = 11.84RR155 pKa = 11.84VTYY158 pKa = 10.49RR159 pKa = 11.84EE160 pKa = 3.9QQKK163 pKa = 9.77EE164 pKa = 4.06RR165 pKa = 11.84SDD167 pKa = 3.74SAVLLTSGCKK177 pKa = 9.46PLPEE181 pKa = 4.38DD182 pKa = 2.88WCLRR186 pKa = 11.84GLGWGGKK193 pKa = 8.16RR194 pKa = 11.84VYY196 pKa = 10.97PMGFWGKK203 pKa = 8.76EE204 pKa = 3.67ANVEE208 pKa = 3.98DD209 pKa = 4.64RR210 pKa = 11.84NVEE213 pKa = 4.03VEE215 pKa = 4.3VLDD218 pKa = 3.96KK219 pKa = 11.68SEE221 pKa = 5.97GGDD224 pKa = 3.3QLDD227 pKa = 5.14GIIEE231 pKa = 4.11DD232 pKa = 3.46EE233 pKa = 4.38RR234 pKa = 11.84EE235 pKa = 3.86EE236 pKa = 4.17NEE238 pKa = 4.24AANEE242 pKa = 4.42LGKK245 pKa = 10.69CWIRR249 pKa = 11.84VACAGLKK256 pKa = 8.95IAKK259 pKa = 9.39HH260 pKa = 5.42VGGFAYY266 pKa = 10.5YY267 pKa = 9.05PAANEE272 pKa = 3.91EE273 pKa = 4.28HH274 pKa = 7.16RR275 pKa = 11.84GQQ277 pKa = 3.8
Molecular weight: 30.64 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5146711 |
12 |
5558 |
459.7 |
50.73 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.52 ± 0.019 | 1.187 ± 0.007 |
5.927 ± 0.015 | 6.181 ± 0.022 |
3.454 ± 0.013 | 6.525 ± 0.02 |
2.547 ± 0.01 | 4.406 ± 0.014 |
4.311 ± 0.022 | 9.077 ± 0.023 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.15 ± 0.009 | 3.098 ± 0.013 |
6.55 ± 0.027 | 3.729 ± 0.014 |
6.594 ± 0.02 | 8.004 ± 0.029 |
5.937 ± 0.016 | 6.606 ± 0.015 |
1.447 ± 0.007 | 2.718 ± 0.012 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
