
Planctomycetes bacterium Poly30
Taxonomy: cellular organisms; Bacteria; PVC group; Planctomycetes; unclassified Planctomycetes
Average proteome isoelectric point is 5.88
Get precalculated fractions of proteins
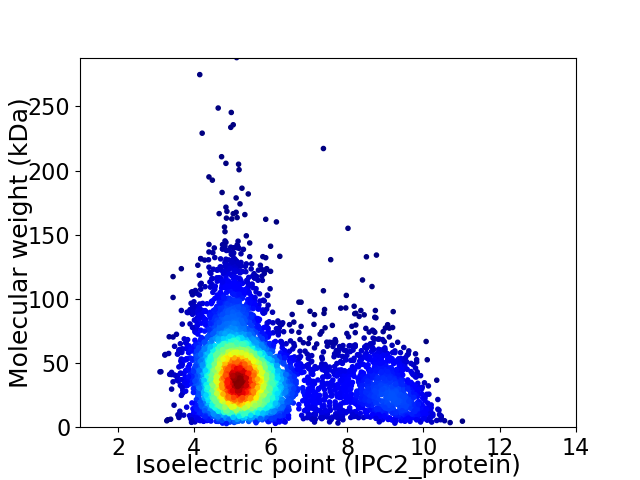
Virtual 2D-PAGE plot for 5628 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
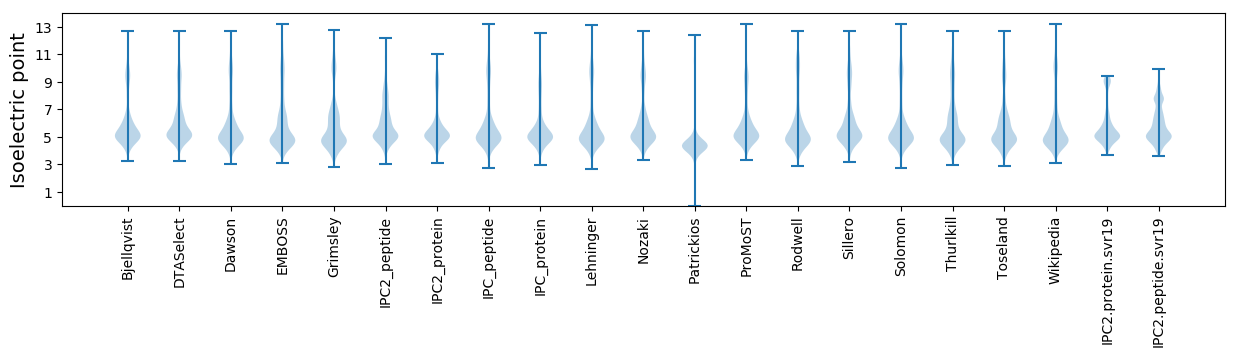
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A518EMG8|A0A518EMG8_9BACT Uncharacterized protein OS=Planctomycetes bacterium Poly30 OX=2528018 GN=Poly30_07810 PE=4 SV=1
MM1 pKa = 7.11QKK3 pKa = 10.53SLLFLAGSALLAIPASAQATFTLMGSGTAADD34 pKa = 4.0MNSDD38 pKa = 2.97GTAVVGRR45 pKa = 11.84DD46 pKa = 3.57GQGAFLWTATAGYY59 pKa = 8.2MPLGGSDD66 pKa = 3.34AVSVSEE72 pKa = 5.24DD73 pKa = 3.29GTVVLGNFTDD83 pKa = 3.76VNGMQSAGRR92 pKa = 11.84WTAATGWVSIGDD104 pKa = 4.1LGSSSGTSLSSAYY117 pKa = 10.27GMSGDD122 pKa = 3.8GNRR125 pKa = 11.84ATGLGWLNAGTAGAFEE141 pKa = 4.42WTPTSGTLQLPQLGPNSSRR160 pKa = 11.84GNCVSLDD167 pKa = 3.33GVYY170 pKa = 10.18IGGWDD175 pKa = 3.94EE176 pKa = 5.65DD177 pKa = 3.77PTGPRR182 pKa = 11.84RR183 pKa = 11.84ASIWGPSGTQFLPLVDD199 pKa = 4.03PVNNPEE205 pKa = 4.51GIGEE209 pKa = 4.27VFGLSSDD216 pKa = 3.18GAYY219 pKa = 10.44AVGGAGDD226 pKa = 3.93TAWLYY231 pKa = 10.46EE232 pKa = 4.26LSTGTLTDD240 pKa = 4.91FGLLPNCCGGFFDD253 pKa = 5.47RR254 pKa = 11.84GFSEE258 pKa = 4.84AVSDD262 pKa = 3.8DD263 pKa = 3.4GQRR266 pKa = 11.84VVGLFGSGPFSFVGTIWGPTSGYY289 pKa = 10.24EE290 pKa = 3.91RR291 pKa = 11.84LSDD294 pKa = 3.63VLVAAGANLQGFEE307 pKa = 4.31ITSGTDD313 pKa = 2.84ISPNGGIILGNAVPTGSFSGQWFIATLDD341 pKa = 3.8PTSGPGTVYY350 pKa = 8.97CTPAAANSTGAPATISTSGSDD371 pKa = 3.26SVANNDD377 pKa = 4.3LVLTASNLPNNSFAYY392 pKa = 9.74FINSMTQAMIVMPGGSQGTVCVGGSVGRR420 pKa = 11.84FLQQIQNSGSNGSISISVDD439 pKa = 3.45LVNLPQPNGLVSVMPGEE456 pKa = 4.31TWNFQAWYY464 pKa = 9.78RR465 pKa = 11.84DD466 pKa = 3.75ANPTSTSNLTDD477 pKa = 5.23AISVTFQQ484 pKa = 2.75
MM1 pKa = 7.11QKK3 pKa = 10.53SLLFLAGSALLAIPASAQATFTLMGSGTAADD34 pKa = 4.0MNSDD38 pKa = 2.97GTAVVGRR45 pKa = 11.84DD46 pKa = 3.57GQGAFLWTATAGYY59 pKa = 8.2MPLGGSDD66 pKa = 3.34AVSVSEE72 pKa = 5.24DD73 pKa = 3.29GTVVLGNFTDD83 pKa = 3.76VNGMQSAGRR92 pKa = 11.84WTAATGWVSIGDD104 pKa = 4.1LGSSSGTSLSSAYY117 pKa = 10.27GMSGDD122 pKa = 3.8GNRR125 pKa = 11.84ATGLGWLNAGTAGAFEE141 pKa = 4.42WTPTSGTLQLPQLGPNSSRR160 pKa = 11.84GNCVSLDD167 pKa = 3.33GVYY170 pKa = 10.18IGGWDD175 pKa = 3.94EE176 pKa = 5.65DD177 pKa = 3.77PTGPRR182 pKa = 11.84RR183 pKa = 11.84ASIWGPSGTQFLPLVDD199 pKa = 4.03PVNNPEE205 pKa = 4.51GIGEE209 pKa = 4.27VFGLSSDD216 pKa = 3.18GAYY219 pKa = 10.44AVGGAGDD226 pKa = 3.93TAWLYY231 pKa = 10.46EE232 pKa = 4.26LSTGTLTDD240 pKa = 4.91FGLLPNCCGGFFDD253 pKa = 5.47RR254 pKa = 11.84GFSEE258 pKa = 4.84AVSDD262 pKa = 3.8DD263 pKa = 3.4GQRR266 pKa = 11.84VVGLFGSGPFSFVGTIWGPTSGYY289 pKa = 10.24EE290 pKa = 3.91RR291 pKa = 11.84LSDD294 pKa = 3.63VLVAAGANLQGFEE307 pKa = 4.31ITSGTDD313 pKa = 2.84ISPNGGIILGNAVPTGSFSGQWFIATLDD341 pKa = 3.8PTSGPGTVYY350 pKa = 8.97CTPAAANSTGAPATISTSGSDD371 pKa = 3.26SVANNDD377 pKa = 4.3LVLTASNLPNNSFAYY392 pKa = 9.74FINSMTQAMIVMPGGSQGTVCVGGSVGRR420 pKa = 11.84FLQQIQNSGSNGSISISVDD439 pKa = 3.45LVNLPQPNGLVSVMPGEE456 pKa = 4.31TWNFQAWYY464 pKa = 9.78RR465 pKa = 11.84DD466 pKa = 3.75ANPTSTSNLTDD477 pKa = 5.23AISVTFQQ484 pKa = 2.75
Molecular weight: 49.08 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A518ESU8|A0A518ESU8_9BACT EamA-like transporter family protein OS=Planctomycetes bacterium Poly30 OX=2528018 GN=Poly30_26880 PE=4 SV=1
MM1 pKa = 7.38KK2 pKa = 10.62LKK4 pKa = 10.4VRR6 pKa = 11.84NSKK9 pKa = 10.29AKK11 pKa = 9.59KK12 pKa = 8.82AKK14 pKa = 8.8KK15 pKa = 8.78TGFLTRR21 pKa = 11.84QKK23 pKa = 10.73SAGGRR28 pKa = 11.84KK29 pKa = 6.56TNKK32 pKa = 9.01RR33 pKa = 11.84QRR35 pKa = 11.84ARR37 pKa = 11.84HH38 pKa = 5.17GSFF41 pKa = 3.22
MM1 pKa = 7.38KK2 pKa = 10.62LKK4 pKa = 10.4VRR6 pKa = 11.84NSKK9 pKa = 10.29AKK11 pKa = 9.59KK12 pKa = 8.82AKK14 pKa = 8.8KK15 pKa = 8.78TGFLTRR21 pKa = 11.84QKK23 pKa = 10.73SAGGRR28 pKa = 11.84KK29 pKa = 6.56TNKK32 pKa = 9.01RR33 pKa = 11.84QRR35 pKa = 11.84ARR37 pKa = 11.84HH38 pKa = 5.17GSFF41 pKa = 3.22
Molecular weight: 4.69 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2299414 |
30 |
2708 |
408.6 |
44.13 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.765 ± 0.044 | 0.824 ± 0.01 |
6.06 ± 0.022 | 6.666 ± 0.039 |
3.588 ± 0.018 | 9.305 ± 0.031 |
2.007 ± 0.013 | 4.005 ± 0.018 |
2.474 ± 0.026 | 10.282 ± 0.037 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.95 ± 0.013 | 2.158 ± 0.022 |
5.604 ± 0.023 | 2.959 ± 0.016 |
7.533 ± 0.039 | 6.447 ± 0.026 |
5.406 ± 0.025 | 7.565 ± 0.025 |
1.472 ± 0.011 | 1.929 ± 0.016 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
