
Enterobacteriaceae bacterium 4M9
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Enterobacterales; Enterobacteriaceae; unclassified Enterobacteriaceae
Average proteome isoelectric point is 6.49
Get precalculated fractions of proteins
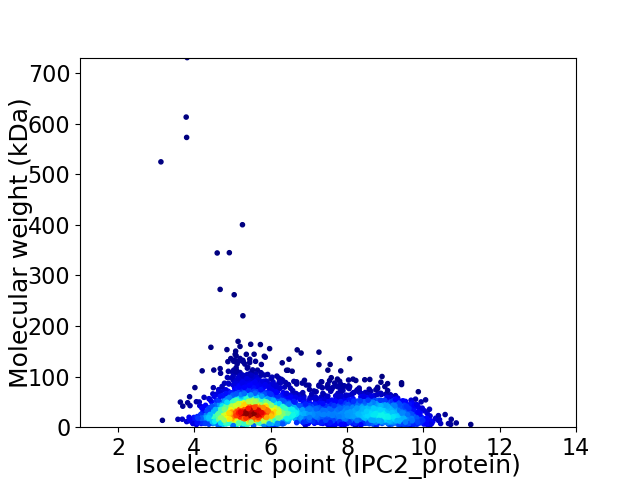
Virtual 2D-PAGE plot for 4055 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
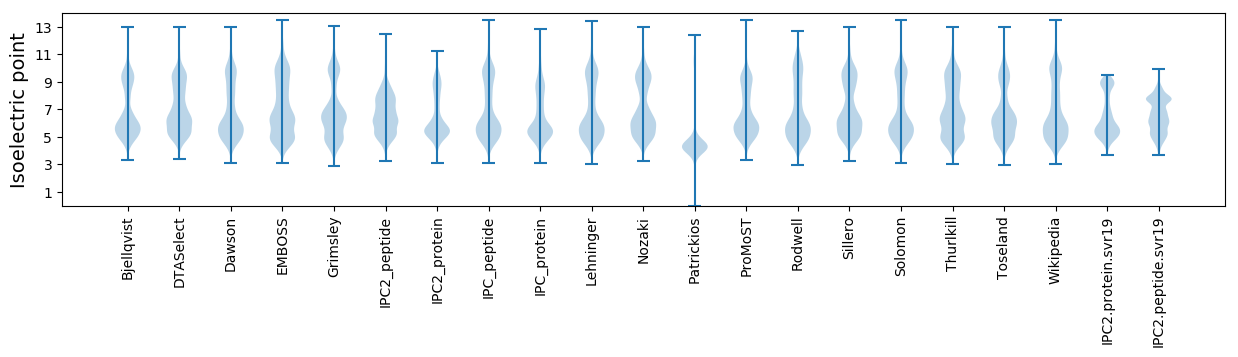
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6B2FKK9|A0A6B2FKK9_9ENTR Flagellar biosynthesis protein FlgN OS=Enterobacteriaceae bacterium 4M9 OX=2699502 GN=GWD52_01605 PE=3 SV=1
MM1 pKa = 7.69SDD3 pKa = 2.89VTLFSHH9 pKa = 6.99RR10 pKa = 11.84GSNPYY15 pKa = 9.38PDD17 pKa = 4.24HH18 pKa = 6.39TLEE21 pKa = 4.86AYY23 pKa = 9.93NAGIDD28 pKa = 3.34WGADD32 pKa = 3.73FIEE35 pKa = 4.99PDD37 pKa = 4.3LYY39 pKa = 9.99ITSDD43 pKa = 3.49GVLVAAHH50 pKa = 7.37DD51 pKa = 4.24DD52 pKa = 3.48MGFANMTYY60 pKa = 10.62AQALAQNSSLMRR72 pKa = 11.84FDD74 pKa = 5.02QIIDD78 pKa = 3.71LVNTRR83 pKa = 11.84SVEE86 pKa = 3.88TGRR89 pKa = 11.84QIGITYY95 pKa = 8.8EE96 pKa = 3.96IKK98 pKa = 9.83NTGNDD103 pKa = 3.53ALTLQAAEE111 pKa = 5.22LSVQMLIDD119 pKa = 3.73KK120 pKa = 10.57GFTNPDD126 pKa = 3.65LIFINSASSDD136 pKa = 3.43TLRR139 pKa = 11.84YY140 pKa = 10.36LNDD143 pKa = 3.12TVMPQSGVDD152 pKa = 3.84FPLIRR157 pKa = 11.84LLSVQEE163 pKa = 4.11SLALKK168 pKa = 10.24YY169 pKa = 10.86GQFNPASVTDD179 pKa = 3.96FADD182 pKa = 5.75GIAVPAYY189 pKa = 8.85IKK191 pKa = 10.95APGVPFSPTINIVDD205 pKa = 3.74SSLVAKK211 pKa = 9.54VHH213 pKa = 5.37AQGLQIHH220 pKa = 5.63TWEE223 pKa = 4.11ALGNEE228 pKa = 4.29FTDD231 pKa = 4.19ARR233 pKa = 11.84LAYY236 pKa = 10.11LDD238 pKa = 3.75KK239 pKa = 10.87TGVDD243 pKa = 4.67AIYY246 pKa = 10.78SDD248 pKa = 5.12YY249 pKa = 10.86IDD251 pKa = 4.13SSTIGLNKK259 pKa = 9.93ADD261 pKa = 3.88GVNTVLGDD269 pKa = 3.89TNSTTLTGTSGDD281 pKa = 3.75DD282 pKa = 3.67VIYY285 pKa = 10.95AMQGDD290 pKa = 4.49DD291 pKa = 3.05IVYY294 pKa = 10.12GKK296 pKa = 10.61GGNDD300 pKa = 3.12VLYY303 pKa = 11.0GDD305 pKa = 5.21GGNDD309 pKa = 3.29TLIAGNGNDD318 pKa = 3.77ILVGGAGNDD327 pKa = 3.39YY328 pKa = 10.71LYY330 pKa = 11.19AGLADD335 pKa = 4.24SLQSQNVLDD344 pKa = 4.32GGVGNDD350 pKa = 3.51VIVTSGTDD358 pKa = 3.25TILYY362 pKa = 7.04GTGDD366 pKa = 4.29GIDD369 pKa = 3.91LLVTNNTSTLQFEE382 pKa = 5.1DD383 pKa = 3.57IARR386 pKa = 11.84QDD388 pKa = 3.26VTFQQYY394 pKa = 11.5NNDD397 pKa = 3.43LVISINNGGALIVQNGASDD416 pKa = 3.85AGSLPGSLVFSDD428 pKa = 6.34GILQTADD435 pKa = 2.91IQTSGSVSPEE445 pKa = 3.87TIAALPDD452 pKa = 3.51LQALLNSGNQLASADD467 pKa = 3.53LWVAA471 pKa = 3.72
MM1 pKa = 7.69SDD3 pKa = 2.89VTLFSHH9 pKa = 6.99RR10 pKa = 11.84GSNPYY15 pKa = 9.38PDD17 pKa = 4.24HH18 pKa = 6.39TLEE21 pKa = 4.86AYY23 pKa = 9.93NAGIDD28 pKa = 3.34WGADD32 pKa = 3.73FIEE35 pKa = 4.99PDD37 pKa = 4.3LYY39 pKa = 9.99ITSDD43 pKa = 3.49GVLVAAHH50 pKa = 7.37DD51 pKa = 4.24DD52 pKa = 3.48MGFANMTYY60 pKa = 10.62AQALAQNSSLMRR72 pKa = 11.84FDD74 pKa = 5.02QIIDD78 pKa = 3.71LVNTRR83 pKa = 11.84SVEE86 pKa = 3.88TGRR89 pKa = 11.84QIGITYY95 pKa = 8.8EE96 pKa = 3.96IKK98 pKa = 9.83NTGNDD103 pKa = 3.53ALTLQAAEE111 pKa = 5.22LSVQMLIDD119 pKa = 3.73KK120 pKa = 10.57GFTNPDD126 pKa = 3.65LIFINSASSDD136 pKa = 3.43TLRR139 pKa = 11.84YY140 pKa = 10.36LNDD143 pKa = 3.12TVMPQSGVDD152 pKa = 3.84FPLIRR157 pKa = 11.84LLSVQEE163 pKa = 4.11SLALKK168 pKa = 10.24YY169 pKa = 10.86GQFNPASVTDD179 pKa = 3.96FADD182 pKa = 5.75GIAVPAYY189 pKa = 8.85IKK191 pKa = 10.95APGVPFSPTINIVDD205 pKa = 3.74SSLVAKK211 pKa = 9.54VHH213 pKa = 5.37AQGLQIHH220 pKa = 5.63TWEE223 pKa = 4.11ALGNEE228 pKa = 4.29FTDD231 pKa = 4.19ARR233 pKa = 11.84LAYY236 pKa = 10.11LDD238 pKa = 3.75KK239 pKa = 10.87TGVDD243 pKa = 4.67AIYY246 pKa = 10.78SDD248 pKa = 5.12YY249 pKa = 10.86IDD251 pKa = 4.13SSTIGLNKK259 pKa = 9.93ADD261 pKa = 3.88GVNTVLGDD269 pKa = 3.89TNSTTLTGTSGDD281 pKa = 3.75DD282 pKa = 3.67VIYY285 pKa = 10.95AMQGDD290 pKa = 4.49DD291 pKa = 3.05IVYY294 pKa = 10.12GKK296 pKa = 10.61GGNDD300 pKa = 3.12VLYY303 pKa = 11.0GDD305 pKa = 5.21GGNDD309 pKa = 3.29TLIAGNGNDD318 pKa = 3.77ILVGGAGNDD327 pKa = 3.39YY328 pKa = 10.71LYY330 pKa = 11.19AGLADD335 pKa = 4.24SLQSQNVLDD344 pKa = 4.32GGVGNDD350 pKa = 3.51VIVTSGTDD358 pKa = 3.25TILYY362 pKa = 7.04GTGDD366 pKa = 4.29GIDD369 pKa = 3.91LLVTNNTSTLQFEE382 pKa = 5.1DD383 pKa = 3.57IARR386 pKa = 11.84QDD388 pKa = 3.26VTFQQYY394 pKa = 11.5NNDD397 pKa = 3.43LVISINNGGALIVQNGASDD416 pKa = 3.85AGSLPGSLVFSDD428 pKa = 6.34GILQTADD435 pKa = 2.91IQTSGSVSPEE445 pKa = 3.87TIAALPDD452 pKa = 3.51LQALLNSGNQLASADD467 pKa = 3.53LWVAA471 pKa = 3.72
Molecular weight: 49.65 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6B2FN82|A0A6B2FN82_9ENTR Uncharacterized MFS-type transporter GWD52_15040 OS=Enterobacteriaceae bacterium 4M9 OX=2699502 GN=GWD52_15040 PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.6RR12 pKa = 11.84NRR14 pKa = 11.84SHH16 pKa = 7.16GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.4NGRR28 pKa = 11.84QVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.22GRR39 pKa = 11.84ARR41 pKa = 11.84LTVSKK46 pKa = 10.99
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.6RR12 pKa = 11.84NRR14 pKa = 11.84SHH16 pKa = 7.16GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.4NGRR28 pKa = 11.84QVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.22GRR39 pKa = 11.84ARR41 pKa = 11.84LTVSKK46 pKa = 10.99
Molecular weight: 5.38 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1319694 |
20 |
7194 |
325.4 |
35.86 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.071 ± 0.044 | 1.067 ± 0.017 |
5.309 ± 0.052 | 5.581 ± 0.04 |
3.717 ± 0.033 | 7.64 ± 0.061 |
2.219 ± 0.019 | 5.428 ± 0.032 |
3.855 ± 0.041 | 10.816 ± 0.073 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.648 ± 0.028 | 3.767 ± 0.04 |
4.471 ± 0.033 | 4.482 ± 0.042 |
5.898 ± 0.06 | 6.069 ± 0.036 |
5.538 ± 0.089 | 7.179 ± 0.04 |
1.549 ± 0.017 | 2.696 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
