
Equus caballus papillomavirus 5
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Dyoiotapapillomavirus; Dyoiotapapillomavirus 2
Average proteome isoelectric point is 6.69
Get precalculated fractions of proteins
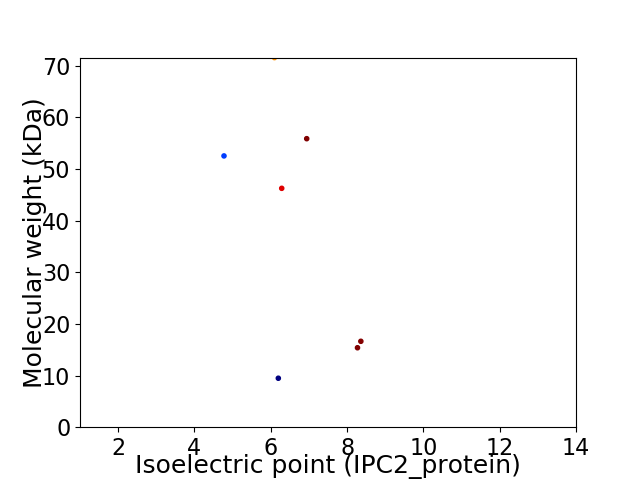
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

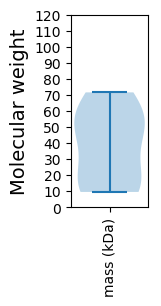
Protein with the lowest isoelectric point:
>tr|K9M8U7|K9M8U7_9PAPI E4 OS=Equus caballus papillomavirus 5 OX=1235429 PE=4 SV=1
MM1 pKa = 8.06DD2 pKa = 5.48APPVKK7 pKa = 9.69RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.37RR11 pKa = 11.84AAVDD15 pKa = 3.33QLYY18 pKa = 7.79KK19 pKa = 7.73TCKK22 pKa = 9.48MGGDD26 pKa = 4.21CPPDD30 pKa = 3.34VVPKK34 pKa = 10.91VEE36 pKa = 4.28GDD38 pKa = 3.71TVADD42 pKa = 6.04RR43 pKa = 11.84ILKK46 pKa = 8.42WLSSFFYY53 pKa = 10.43FGNLGISTGRR63 pKa = 11.84GTGGRR68 pKa = 11.84LGYY71 pKa = 8.94TPLGGGGGHH80 pKa = 6.55AAEE83 pKa = 4.93GGGVRR88 pKa = 11.84VGAPISTLGVGVDD101 pKa = 3.56AVGPAEE107 pKa = 4.25AVPVDD112 pKa = 3.9VLQPSAPSIIPLGEE126 pKa = 4.12GVSVDD131 pKa = 3.64TVDD134 pKa = 4.68VIAEE138 pKa = 4.07VLPPPGGARR147 pKa = 11.84PEE149 pKa = 4.11VTAEE153 pKa = 4.16VPGQPATIDD162 pKa = 3.26VAMDD166 pKa = 3.07VVPRR170 pKa = 11.84LRR172 pKa = 11.84AAVSRR177 pKa = 11.84STFHH181 pKa = 6.67NPAFQVEE188 pKa = 4.43LQSLGTGEE196 pKa = 4.44SSASEE201 pKa = 3.65QAFVFGHH208 pKa = 5.8AGGRR212 pKa = 11.84LVDD215 pKa = 3.89TVGEE219 pKa = 4.49DD220 pKa = 3.21IEE222 pKa = 4.65MVALGDD228 pKa = 3.84GVPRR232 pKa = 11.84TSTPRR237 pKa = 11.84SSSATPRR244 pKa = 11.84SRR246 pKa = 11.84LWGRR250 pKa = 11.84RR251 pKa = 11.84FEE253 pKa = 4.26QVQVADD259 pKa = 4.18PAFLSAPDD267 pKa = 3.71TLVQYY272 pKa = 10.23GFSNPAYY279 pKa = 9.95DD280 pKa = 4.17PEE282 pKa = 6.07ASLVFPYY289 pKa = 10.61TEE291 pKa = 4.32GEE293 pKa = 3.81ARR295 pKa = 11.84AAPSTLFQDD304 pKa = 3.47VAYY307 pKa = 10.42LGRR310 pKa = 11.84PEE312 pKa = 3.97ITSHH316 pKa = 6.45KK317 pKa = 9.85EE318 pKa = 3.71SIGVSRR324 pKa = 11.84LGRR327 pKa = 11.84RR328 pKa = 11.84ATVQTRR334 pKa = 11.84SGTIIGAEE342 pKa = 3.65VHH344 pKa = 6.25FRR346 pKa = 11.84YY347 pKa = 9.68EE348 pKa = 4.04LSPITSEE355 pKa = 5.02DD356 pKa = 3.23IEE358 pKa = 4.92LSVLTTQDD366 pKa = 3.22EE367 pKa = 4.51EE368 pKa = 4.36ALSVVDD374 pKa = 5.88LEE376 pKa = 4.96SLTSAYY382 pKa = 9.65SEE384 pKa = 4.42RR385 pKa = 11.84EE386 pKa = 4.06LLEE389 pKa = 5.27DD390 pKa = 4.29DD391 pKa = 3.84QPNIHH396 pKa = 6.36GVLSFTDD403 pKa = 3.62EE404 pKa = 4.2EE405 pKa = 5.08GATTSLPLAPPQARR419 pKa = 11.84FFYY422 pKa = 10.15TGPFLQGGRR431 pKa = 11.84GVSDD435 pKa = 3.42GRR437 pKa = 11.84GQAAGEE443 pKa = 4.24SSHH446 pKa = 7.22PGTDD450 pKa = 3.45VIIEE454 pKa = 4.05YY455 pKa = 10.06PEE457 pKa = 5.0AGGSYY462 pKa = 9.76FLHH465 pKa = 6.37PTAPCRR471 pKa = 11.84RR472 pKa = 11.84KK473 pKa = 9.94RR474 pKa = 11.84RR475 pKa = 11.84YY476 pKa = 9.9CFADD480 pKa = 3.62GLLDD484 pKa = 4.31AGQSEE489 pKa = 5.1VLSPPCACYY498 pKa = 10.36
MM1 pKa = 8.06DD2 pKa = 5.48APPVKK7 pKa = 9.69RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.37RR11 pKa = 11.84AAVDD15 pKa = 3.33QLYY18 pKa = 7.79KK19 pKa = 7.73TCKK22 pKa = 9.48MGGDD26 pKa = 4.21CPPDD30 pKa = 3.34VVPKK34 pKa = 10.91VEE36 pKa = 4.28GDD38 pKa = 3.71TVADD42 pKa = 6.04RR43 pKa = 11.84ILKK46 pKa = 8.42WLSSFFYY53 pKa = 10.43FGNLGISTGRR63 pKa = 11.84GTGGRR68 pKa = 11.84LGYY71 pKa = 8.94TPLGGGGGHH80 pKa = 6.55AAEE83 pKa = 4.93GGGVRR88 pKa = 11.84VGAPISTLGVGVDD101 pKa = 3.56AVGPAEE107 pKa = 4.25AVPVDD112 pKa = 3.9VLQPSAPSIIPLGEE126 pKa = 4.12GVSVDD131 pKa = 3.64TVDD134 pKa = 4.68VIAEE138 pKa = 4.07VLPPPGGARR147 pKa = 11.84PEE149 pKa = 4.11VTAEE153 pKa = 4.16VPGQPATIDD162 pKa = 3.26VAMDD166 pKa = 3.07VVPRR170 pKa = 11.84LRR172 pKa = 11.84AAVSRR177 pKa = 11.84STFHH181 pKa = 6.67NPAFQVEE188 pKa = 4.43LQSLGTGEE196 pKa = 4.44SSASEE201 pKa = 3.65QAFVFGHH208 pKa = 5.8AGGRR212 pKa = 11.84LVDD215 pKa = 3.89TVGEE219 pKa = 4.49DD220 pKa = 3.21IEE222 pKa = 4.65MVALGDD228 pKa = 3.84GVPRR232 pKa = 11.84TSTPRR237 pKa = 11.84SSSATPRR244 pKa = 11.84SRR246 pKa = 11.84LWGRR250 pKa = 11.84RR251 pKa = 11.84FEE253 pKa = 4.26QVQVADD259 pKa = 4.18PAFLSAPDD267 pKa = 3.71TLVQYY272 pKa = 10.23GFSNPAYY279 pKa = 9.95DD280 pKa = 4.17PEE282 pKa = 6.07ASLVFPYY289 pKa = 10.61TEE291 pKa = 4.32GEE293 pKa = 3.81ARR295 pKa = 11.84AAPSTLFQDD304 pKa = 3.47VAYY307 pKa = 10.42LGRR310 pKa = 11.84PEE312 pKa = 3.97ITSHH316 pKa = 6.45KK317 pKa = 9.85EE318 pKa = 3.71SIGVSRR324 pKa = 11.84LGRR327 pKa = 11.84RR328 pKa = 11.84ATVQTRR334 pKa = 11.84SGTIIGAEE342 pKa = 3.65VHH344 pKa = 6.25FRR346 pKa = 11.84YY347 pKa = 9.68EE348 pKa = 4.04LSPITSEE355 pKa = 5.02DD356 pKa = 3.23IEE358 pKa = 4.92LSVLTTQDD366 pKa = 3.22EE367 pKa = 4.51EE368 pKa = 4.36ALSVVDD374 pKa = 5.88LEE376 pKa = 4.96SLTSAYY382 pKa = 9.65SEE384 pKa = 4.42RR385 pKa = 11.84EE386 pKa = 4.06LLEE389 pKa = 5.27DD390 pKa = 4.29DD391 pKa = 3.84QPNIHH396 pKa = 6.36GVLSFTDD403 pKa = 3.62EE404 pKa = 4.2EE405 pKa = 5.08GATTSLPLAPPQARR419 pKa = 11.84FFYY422 pKa = 10.15TGPFLQGGRR431 pKa = 11.84GVSDD435 pKa = 3.42GRR437 pKa = 11.84GQAAGEE443 pKa = 4.24SSHH446 pKa = 7.22PGTDD450 pKa = 3.45VIIEE454 pKa = 4.05YY455 pKa = 10.06PEE457 pKa = 5.0AGGSYY462 pKa = 9.76FLHH465 pKa = 6.37PTAPCRR471 pKa = 11.84RR472 pKa = 11.84KK473 pKa = 9.94RR474 pKa = 11.84RR475 pKa = 11.84YY476 pKa = 9.9CFADD480 pKa = 3.62GLLDD484 pKa = 4.31AGQSEE489 pKa = 5.1VLSPPCACYY498 pKa = 10.36
Molecular weight: 52.51 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|K9M934|K9M934_9PAPI E7 OS=Equus caballus papillomavirus 5 OX=1235429 PE=4 SV=1
MM1 pKa = 7.36KK2 pKa = 9.89PSPPLLSPAQVVVTARR18 pKa = 11.84KK19 pKa = 9.54EE20 pKa = 3.92EE21 pKa = 4.25AGTDD25 pKa = 3.67QTDD28 pKa = 3.49SAATQAQGGLAQFPEE43 pKa = 4.33TLVDD47 pKa = 3.58YY48 pKa = 10.46LKK50 pKa = 10.86RR51 pKa = 11.84NHH53 pKa = 6.25LTGADD58 pKa = 3.21VWPRR62 pKa = 11.84QSGCLLNSRR71 pKa = 11.84GKK73 pKa = 10.55ALLQCMTAGTAEE85 pKa = 4.18EE86 pKa = 4.63APLAVTPSLASEE98 pKa = 3.78ARR100 pKa = 11.84GYY102 pKa = 9.87WEE104 pKa = 5.05IPPPPAPCRR113 pKa = 11.84ARR115 pKa = 11.84MRR117 pKa = 11.84RR118 pKa = 11.84TRR120 pKa = 11.84LAHH123 pKa = 5.66RR124 pKa = 11.84QRR126 pKa = 11.84LKK128 pKa = 8.98QTLWHH133 pKa = 6.3LQRR136 pKa = 11.84EE137 pKa = 4.62LSEE140 pKa = 3.8LMYY143 pKa = 10.74WMDD146 pKa = 3.92VGILL150 pKa = 3.51
MM1 pKa = 7.36KK2 pKa = 9.89PSPPLLSPAQVVVTARR18 pKa = 11.84KK19 pKa = 9.54EE20 pKa = 3.92EE21 pKa = 4.25AGTDD25 pKa = 3.67QTDD28 pKa = 3.49SAATQAQGGLAQFPEE43 pKa = 4.33TLVDD47 pKa = 3.58YY48 pKa = 10.46LKK50 pKa = 10.86RR51 pKa = 11.84NHH53 pKa = 6.25LTGADD58 pKa = 3.21VWPRR62 pKa = 11.84QSGCLLNSRR71 pKa = 11.84GKK73 pKa = 10.55ALLQCMTAGTAEE85 pKa = 4.18EE86 pKa = 4.63APLAVTPSLASEE98 pKa = 3.78ARR100 pKa = 11.84GYY102 pKa = 9.87WEE104 pKa = 5.05IPPPPAPCRR113 pKa = 11.84ARR115 pKa = 11.84MRR117 pKa = 11.84RR118 pKa = 11.84TRR120 pKa = 11.84LAHH123 pKa = 5.66RR124 pKa = 11.84QRR126 pKa = 11.84LKK128 pKa = 8.98QTLWHH133 pKa = 6.3LQRR136 pKa = 11.84EE137 pKa = 4.62LSEE140 pKa = 3.8LMYY143 pKa = 10.74WMDD146 pKa = 3.92VGILL150 pKa = 3.51
Molecular weight: 16.63 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2411 |
84 |
632 |
344.4 |
38.24 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.715 ± 0.88 | 2.406 ± 0.57 |
5.89 ± 0.42 | 5.931 ± 0.216 |
4.106 ± 0.57 | 8.295 ± 1.13 |
1.742 ± 0.191 | 2.986 ± 0.541 |
4.148 ± 0.608 | 8.337 ± 0.45 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.701 ± 0.327 | 3.36 ± 0.892 |
6.802 ± 0.528 | 4.148 ± 0.26 |
7.258 ± 0.414 | 7.507 ± 0.604 |
5.641 ± 0.398 | 6.802 ± 0.539 |
1.866 ± 0.395 | 3.36 ± 0.432 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
