
Hubei sobemo-like virus 43
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.13
Get precalculated fractions of proteins
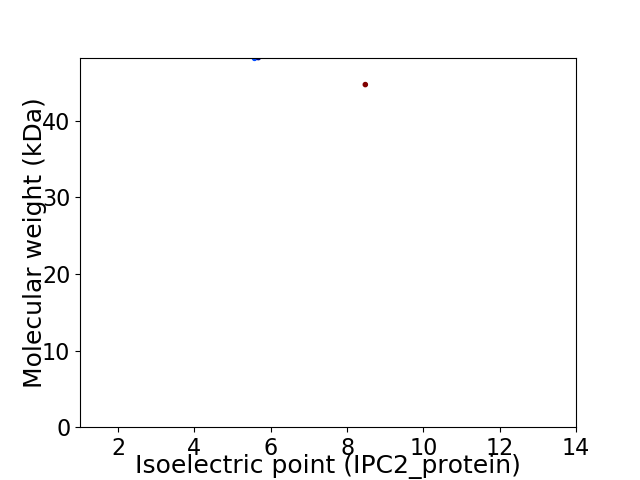
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KE23|A0A1L3KE23_9VIRU RNA-directed RNA polymerase OS=Hubei sobemo-like virus 43 OX=1923231 PE=4 SV=1
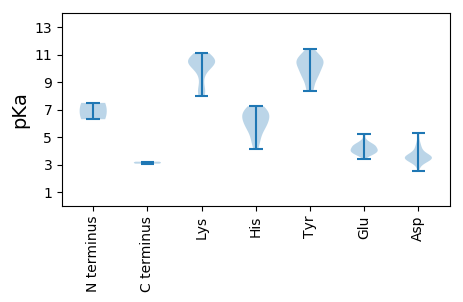
MM1 pKa = 7.48PRR3 pKa = 11.84GGAVDD8 pKa = 3.73EE9 pKa = 4.32LASLKK14 pKa = 10.67AHH16 pKa = 6.76AEE18 pKa = 3.97FFRR21 pKa = 11.84QSRR24 pKa = 11.84EE25 pKa = 3.71SGLEE29 pKa = 3.71VEE31 pKa = 4.43PLDD34 pKa = 4.97RR35 pKa = 11.84EE36 pKa = 4.51RR37 pKa = 11.84ILHH40 pKa = 5.65ACEE43 pKa = 3.37TAYY46 pKa = 10.62AAARR50 pKa = 11.84YY51 pKa = 8.55SLPSDD56 pKa = 3.18WLEE59 pKa = 3.67KK60 pKa = 7.99THH62 pKa = 7.15FYY64 pKa = 10.68RR65 pKa = 11.84VVYY68 pKa = 10.17DD69 pKa = 3.44LEE71 pKa = 4.62FRR73 pKa = 11.84SSPGYY78 pKa = 9.21PYY80 pKa = 10.83CLASSTIGDD89 pKa = 3.53WLGWNGVGCDD99 pKa = 3.78GEE101 pKa = 4.42KK102 pKa = 10.88LEE104 pKa = 4.7EE105 pKa = 3.93LWFDD109 pKa = 3.46VLAFLSGEE117 pKa = 3.67RR118 pKa = 11.84DD119 pKa = 2.9SFYY122 pKa = 11.06RR123 pKa = 11.84VFIKK127 pKa = 10.6SEE129 pKa = 3.68PHH131 pKa = 4.97TLAKK135 pKa = 9.96RR136 pKa = 11.84DD137 pKa = 3.45AGRR140 pKa = 11.84WRR142 pKa = 11.84LIICPPLYY150 pKa = 10.51EE151 pKa = 4.04QVAWTMVFGPGNDD164 pKa = 3.44RR165 pKa = 11.84EE166 pKa = 4.29IEE168 pKa = 4.48TVGRR172 pKa = 11.84TPSMQGMSLPGGLWKK187 pKa = 10.54DD188 pKa = 3.15WLAIFQQRR196 pKa = 11.84GLNVAMDD203 pKa = 3.96KK204 pKa = 11.11SAWDD208 pKa = 3.55WTAHH212 pKa = 4.9EE213 pKa = 4.63QLISMDD219 pKa = 3.28LEE221 pKa = 4.15LRR223 pKa = 11.84HH224 pKa = 6.55RR225 pKa = 11.84LLTGPNRR232 pKa = 11.84GRR234 pKa = 11.84WRR236 pKa = 11.84QLAEE240 pKa = 3.86RR241 pKa = 11.84LYY243 pKa = 11.11DD244 pKa = 3.44GAFNHH249 pKa = 6.46PRR251 pKa = 11.84LVLSSGAVYY260 pKa = 10.1QQTEE264 pKa = 4.22PGVMKK269 pKa = 10.25SGCVNTISSNSHH281 pKa = 4.15MQVFVHH287 pKa = 6.68CLACLRR293 pKa = 11.84YY294 pKa = 9.4GLPLTPLPVAVGDD307 pKa = 4.13DD308 pKa = 3.99TLSSLSNLVEE318 pKa = 3.72PSAYY322 pKa = 9.93RR323 pKa = 11.84FTGAVVKK330 pKa = 10.47EE331 pKa = 4.29VDD333 pKa = 3.62LDD335 pKa = 3.55MHH337 pKa = 6.34FVGHH341 pKa = 6.54RR342 pKa = 11.84WYY344 pKa = 10.33EE345 pKa = 4.07SGPVPTYY352 pKa = 9.63SAKK355 pKa = 10.36HH356 pKa = 4.56FVRR359 pKa = 11.84FAVTGADD366 pKa = 3.69YY367 pKa = 11.42VPDD370 pKa = 4.3FLDD373 pKa = 3.81SMVRR377 pKa = 11.84LYY379 pKa = 11.02AHH381 pKa = 7.05HH382 pKa = 6.99EE383 pKa = 4.26GFQKK387 pKa = 9.49VWRR390 pKa = 11.84WLAYY394 pKa = 9.91RR395 pKa = 11.84RR396 pKa = 11.84GIDD399 pKa = 3.53LPSEE403 pKa = 4.08AFVKK407 pKa = 10.43FWYY410 pKa = 9.64DD411 pKa = 3.35YY412 pKa = 10.47PDD414 pKa = 5.31DD415 pKa = 4.61VIQYY419 pKa = 9.62YY420 pKa = 10.36GG421 pKa = 3.21
MM1 pKa = 7.48PRR3 pKa = 11.84GGAVDD8 pKa = 3.73EE9 pKa = 4.32LASLKK14 pKa = 10.67AHH16 pKa = 6.76AEE18 pKa = 3.97FFRR21 pKa = 11.84QSRR24 pKa = 11.84EE25 pKa = 3.71SGLEE29 pKa = 3.71VEE31 pKa = 4.43PLDD34 pKa = 4.97RR35 pKa = 11.84EE36 pKa = 4.51RR37 pKa = 11.84ILHH40 pKa = 5.65ACEE43 pKa = 3.37TAYY46 pKa = 10.62AAARR50 pKa = 11.84YY51 pKa = 8.55SLPSDD56 pKa = 3.18WLEE59 pKa = 3.67KK60 pKa = 7.99THH62 pKa = 7.15FYY64 pKa = 10.68RR65 pKa = 11.84VVYY68 pKa = 10.17DD69 pKa = 3.44LEE71 pKa = 4.62FRR73 pKa = 11.84SSPGYY78 pKa = 9.21PYY80 pKa = 10.83CLASSTIGDD89 pKa = 3.53WLGWNGVGCDD99 pKa = 3.78GEE101 pKa = 4.42KK102 pKa = 10.88LEE104 pKa = 4.7EE105 pKa = 3.93LWFDD109 pKa = 3.46VLAFLSGEE117 pKa = 3.67RR118 pKa = 11.84DD119 pKa = 2.9SFYY122 pKa = 11.06RR123 pKa = 11.84VFIKK127 pKa = 10.6SEE129 pKa = 3.68PHH131 pKa = 4.97TLAKK135 pKa = 9.96RR136 pKa = 11.84DD137 pKa = 3.45AGRR140 pKa = 11.84WRR142 pKa = 11.84LIICPPLYY150 pKa = 10.51EE151 pKa = 4.04QVAWTMVFGPGNDD164 pKa = 3.44RR165 pKa = 11.84EE166 pKa = 4.29IEE168 pKa = 4.48TVGRR172 pKa = 11.84TPSMQGMSLPGGLWKK187 pKa = 10.54DD188 pKa = 3.15WLAIFQQRR196 pKa = 11.84GLNVAMDD203 pKa = 3.96KK204 pKa = 11.11SAWDD208 pKa = 3.55WTAHH212 pKa = 4.9EE213 pKa = 4.63QLISMDD219 pKa = 3.28LEE221 pKa = 4.15LRR223 pKa = 11.84HH224 pKa = 6.55RR225 pKa = 11.84LLTGPNRR232 pKa = 11.84GRR234 pKa = 11.84WRR236 pKa = 11.84QLAEE240 pKa = 3.86RR241 pKa = 11.84LYY243 pKa = 11.11DD244 pKa = 3.44GAFNHH249 pKa = 6.46PRR251 pKa = 11.84LVLSSGAVYY260 pKa = 10.1QQTEE264 pKa = 4.22PGVMKK269 pKa = 10.25SGCVNTISSNSHH281 pKa = 4.15MQVFVHH287 pKa = 6.68CLACLRR293 pKa = 11.84YY294 pKa = 9.4GLPLTPLPVAVGDD307 pKa = 4.13DD308 pKa = 3.99TLSSLSNLVEE318 pKa = 3.72PSAYY322 pKa = 9.93RR323 pKa = 11.84FTGAVVKK330 pKa = 10.47EE331 pKa = 4.29VDD333 pKa = 3.62LDD335 pKa = 3.55MHH337 pKa = 6.34FVGHH341 pKa = 6.54RR342 pKa = 11.84WYY344 pKa = 10.33EE345 pKa = 4.07SGPVPTYY352 pKa = 9.63SAKK355 pKa = 10.36HH356 pKa = 4.56FVRR359 pKa = 11.84FAVTGADD366 pKa = 3.69YY367 pKa = 11.42VPDD370 pKa = 4.3FLDD373 pKa = 3.81SMVRR377 pKa = 11.84LYY379 pKa = 11.02AHH381 pKa = 7.05HH382 pKa = 6.99EE383 pKa = 4.26GFQKK387 pKa = 9.49VWRR390 pKa = 11.84WLAYY394 pKa = 9.91RR395 pKa = 11.84RR396 pKa = 11.84GIDD399 pKa = 3.53LPSEE403 pKa = 4.08AFVKK407 pKa = 10.43FWYY410 pKa = 9.64DD411 pKa = 3.35YY412 pKa = 10.47PDD414 pKa = 5.31DD415 pKa = 4.61VIQYY419 pKa = 9.62YY420 pKa = 10.36GG421 pKa = 3.21
Molecular weight: 48.15 kDa
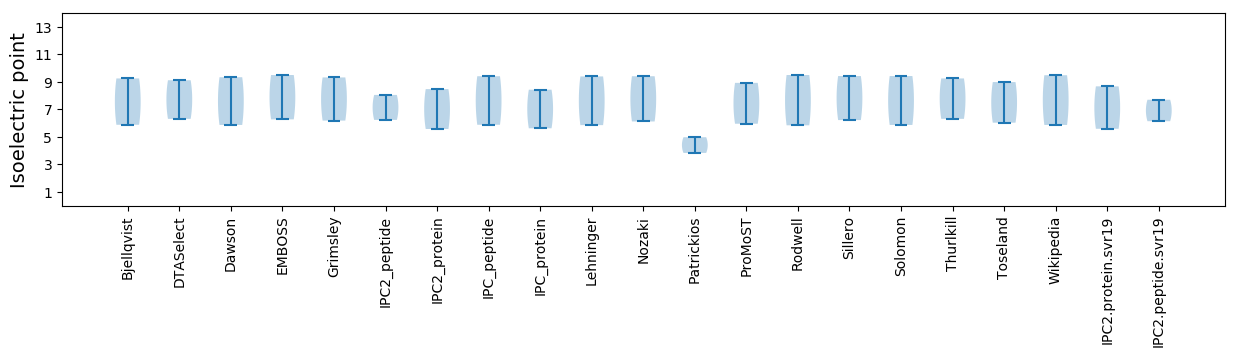
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KE23|A0A1L3KE23_9VIRU RNA-directed RNA polymerase OS=Hubei sobemo-like virus 43 OX=1923231 PE=4 SV=1
MM1 pKa = 6.3TTASFMFALARR12 pKa = 11.84KK13 pKa = 8.29QLPTAPPNPGWGIGLARR30 pKa = 11.84PTLRR34 pKa = 11.84EE35 pKa = 3.76GLVLVGGVAATAASGYY51 pKa = 10.3AVYY54 pKa = 10.61SVGCWTMSACRR65 pKa = 11.84RR66 pKa = 11.84LRR68 pKa = 11.84SWWAKK73 pKa = 8.46PLKK76 pKa = 10.11LRR78 pKa = 11.84PAPSGPVVGEE88 pKa = 4.18CVPEE92 pKa = 4.13SAVPGSEE99 pKa = 4.38EE100 pKa = 5.18RR101 pKa = 11.84PMTPPKK107 pKa = 10.45GQALVGFSDD116 pKa = 4.53GSSFMVVGCAVRR128 pKa = 11.84MEE130 pKa = 4.81DD131 pKa = 3.31WLVMPDD137 pKa = 3.51HH138 pKa = 6.87VKK140 pKa = 10.6SALGDD145 pKa = 3.23RR146 pKa = 11.84RR147 pKa = 11.84LEE149 pKa = 3.98IRR151 pKa = 11.84SMDD154 pKa = 3.59HH155 pKa = 6.13KK156 pKa = 10.89RR157 pKa = 11.84VVTLHH162 pKa = 5.53NAEE165 pKa = 4.13VEE167 pKa = 4.04AMQLVDD173 pKa = 3.56TDD175 pKa = 4.34LLAVQLSPARR185 pKa = 11.84FSEE188 pKa = 3.84IGLAKK193 pKa = 9.64VTVGPNLAEE202 pKa = 4.01KK203 pKa = 10.35FGAFSAIVGAFGKK216 pKa = 9.0GTTGTVKK223 pKa = 10.27HH224 pKa = 5.98AQLFGKK230 pKa = 8.15ITYY233 pKa = 8.39TGSTFKK239 pKa = 10.63GYY241 pKa = 10.62SGAAYY246 pKa = 8.37MAGTQLVGIHH256 pKa = 5.68LHH258 pKa = 6.27GGTVNAGYY266 pKa = 9.72SASYY270 pKa = 9.02VLAMLKK276 pKa = 10.6HH277 pKa = 5.6MFRR280 pKa = 11.84IKK282 pKa = 10.91DD283 pKa = 3.46EE284 pKa = 5.24GSDD287 pKa = 3.23EE288 pKa = 4.03WLEE291 pKa = 3.9GMRR294 pKa = 11.84RR295 pKa = 11.84QGAEE299 pKa = 3.92VVVDD303 pKa = 3.59QDD305 pKa = 3.03WRR307 pKa = 11.84DD308 pKa = 3.24MDD310 pKa = 3.62EE311 pKa = 3.99CRR313 pKa = 11.84IRR315 pKa = 11.84VAGRR319 pKa = 11.84YY320 pKa = 9.18HH321 pKa = 7.23IIGVDD326 pKa = 3.3TMSRR330 pKa = 11.84VYY332 pKa = 10.58GADD335 pKa = 2.56WRR337 pKa = 11.84RR338 pKa = 11.84SGKK341 pKa = 10.32RR342 pKa = 11.84NLKK345 pKa = 8.31TDD347 pKa = 3.48RR348 pKa = 11.84FVDD351 pKa = 4.1LEE353 pKa = 4.46SCVLPGEE360 pKa = 4.34SSEE363 pKa = 4.29TLSGGSRR370 pKa = 11.84VSVPSNPSLAEE381 pKa = 3.81LANQLTLHH389 pKa = 5.99EE390 pKa = 4.42LEE392 pKa = 4.46KK393 pKa = 10.89LRR395 pKa = 11.84EE396 pKa = 3.9RR397 pKa = 11.84LALRR401 pKa = 11.84AKK403 pKa = 10.34DD404 pKa = 3.71LRR406 pKa = 11.84ASATGSRR413 pKa = 11.84AA414 pKa = 3.06
MM1 pKa = 6.3TTASFMFALARR12 pKa = 11.84KK13 pKa = 8.29QLPTAPPNPGWGIGLARR30 pKa = 11.84PTLRR34 pKa = 11.84EE35 pKa = 3.76GLVLVGGVAATAASGYY51 pKa = 10.3AVYY54 pKa = 10.61SVGCWTMSACRR65 pKa = 11.84RR66 pKa = 11.84LRR68 pKa = 11.84SWWAKK73 pKa = 8.46PLKK76 pKa = 10.11LRR78 pKa = 11.84PAPSGPVVGEE88 pKa = 4.18CVPEE92 pKa = 4.13SAVPGSEE99 pKa = 4.38EE100 pKa = 5.18RR101 pKa = 11.84PMTPPKK107 pKa = 10.45GQALVGFSDD116 pKa = 4.53GSSFMVVGCAVRR128 pKa = 11.84MEE130 pKa = 4.81DD131 pKa = 3.31WLVMPDD137 pKa = 3.51HH138 pKa = 6.87VKK140 pKa = 10.6SALGDD145 pKa = 3.23RR146 pKa = 11.84RR147 pKa = 11.84LEE149 pKa = 3.98IRR151 pKa = 11.84SMDD154 pKa = 3.59HH155 pKa = 6.13KK156 pKa = 10.89RR157 pKa = 11.84VVTLHH162 pKa = 5.53NAEE165 pKa = 4.13VEE167 pKa = 4.04AMQLVDD173 pKa = 3.56TDD175 pKa = 4.34LLAVQLSPARR185 pKa = 11.84FSEE188 pKa = 3.84IGLAKK193 pKa = 9.64VTVGPNLAEE202 pKa = 4.01KK203 pKa = 10.35FGAFSAIVGAFGKK216 pKa = 9.0GTTGTVKK223 pKa = 10.27HH224 pKa = 5.98AQLFGKK230 pKa = 8.15ITYY233 pKa = 8.39TGSTFKK239 pKa = 10.63GYY241 pKa = 10.62SGAAYY246 pKa = 8.37MAGTQLVGIHH256 pKa = 5.68LHH258 pKa = 6.27GGTVNAGYY266 pKa = 9.72SASYY270 pKa = 9.02VLAMLKK276 pKa = 10.6HH277 pKa = 5.6MFRR280 pKa = 11.84IKK282 pKa = 10.91DD283 pKa = 3.46EE284 pKa = 5.24GSDD287 pKa = 3.23EE288 pKa = 4.03WLEE291 pKa = 3.9GMRR294 pKa = 11.84RR295 pKa = 11.84QGAEE299 pKa = 3.92VVVDD303 pKa = 3.59QDD305 pKa = 3.03WRR307 pKa = 11.84DD308 pKa = 3.24MDD310 pKa = 3.62EE311 pKa = 3.99CRR313 pKa = 11.84IRR315 pKa = 11.84VAGRR319 pKa = 11.84YY320 pKa = 9.18HH321 pKa = 7.23IIGVDD326 pKa = 3.3TMSRR330 pKa = 11.84VYY332 pKa = 10.58GADD335 pKa = 2.56WRR337 pKa = 11.84RR338 pKa = 11.84SGKK341 pKa = 10.32RR342 pKa = 11.84NLKK345 pKa = 8.31TDD347 pKa = 3.48RR348 pKa = 11.84FVDD351 pKa = 4.1LEE353 pKa = 4.46SCVLPGEE360 pKa = 4.34SSEE363 pKa = 4.29TLSGGSRR370 pKa = 11.84VSVPSNPSLAEE381 pKa = 3.81LANQLTLHH389 pKa = 5.99EE390 pKa = 4.42LEE392 pKa = 4.46KK393 pKa = 10.89LRR395 pKa = 11.84EE396 pKa = 3.9RR397 pKa = 11.84LALRR401 pKa = 11.84AKK403 pKa = 10.34DD404 pKa = 3.71LRR406 pKa = 11.84ASATGSRR413 pKa = 11.84AA414 pKa = 3.06
Molecular weight: 44.73 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
835 |
414 |
421 |
417.5 |
46.44 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.862 ± 1.127 | 1.557 ± 0.08 |
5.269 ± 0.681 | 5.868 ± 0.231 |
3.832 ± 0.69 | 9.102 ± 0.95 |
2.754 ± 0.429 | 2.515 ± 0.074 |
3.593 ± 0.558 | 9.82 ± 0.296 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.994 ± 0.465 | 1.796 ± 0.078 |
5.15 ± 0.236 | 2.515 ± 0.252 |
7.545 ± 0.136 | 7.545 ± 0.315 |
4.671 ± 0.654 | 8.263 ± 0.498 |
2.754 ± 0.608 | 3.593 ± 1.049 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
