
Donghicola tyrosinivorans
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Roseobacteraceae; Donghicola
Average proteome isoelectric point is 6.07
Get precalculated fractions of proteins
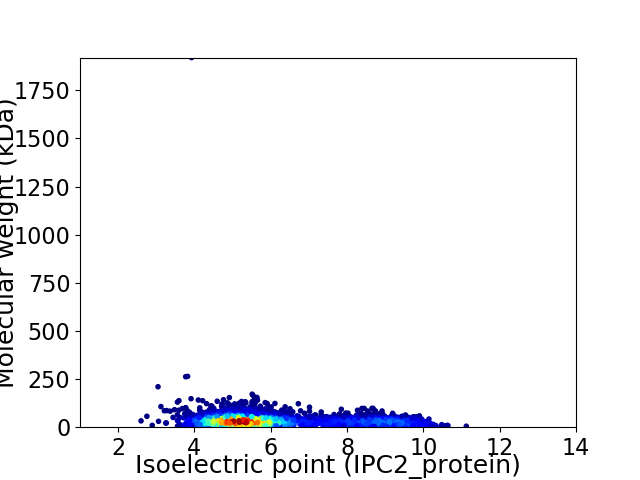
Virtual 2D-PAGE plot for 4472 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2T0W7T4|A0A2T0W7T4_9RHOB Uncharacterized protein OS=Donghicola tyrosinivorans OX=1652492 GN=CLV74_1391 PE=4 SV=1
MM1 pKa = 7.14ATSTTFQVMYY11 pKa = 10.02LGQRR15 pKa = 11.84ALIDD19 pKa = 3.42TTQGNNTAEE28 pKa = 4.12NAAGILDD35 pKa = 4.34TYY37 pKa = 11.34GSASDD42 pKa = 4.76PMYY45 pKa = 10.83NWVGTLYY52 pKa = 10.99ADD54 pKa = 4.74YY55 pKa = 11.14LLEE58 pKa = 5.5DD59 pKa = 5.18DD60 pKa = 4.68NNSYY64 pKa = 11.11DD65 pKa = 3.35IDD67 pKa = 3.93NGGGRR72 pKa = 11.84DD73 pKa = 3.26RR74 pKa = 11.84FHH76 pKa = 8.1VNGFGTQEE84 pKa = 3.43FDD86 pKa = 3.89AVAEE90 pKa = 4.07YY91 pKa = 10.35AITLTYY97 pKa = 10.88VDD99 pKa = 4.32GTTAQITAYY108 pKa = 10.27VFQDD112 pKa = 3.76FQGHH116 pKa = 5.28TFLAPEE122 pKa = 4.17TTLNADD128 pKa = 3.49QAALTAKK135 pKa = 9.94PIQSLALTAVTKK147 pKa = 10.84SSGDD151 pKa = 3.39TTAGDD156 pKa = 4.08MIAARR161 pKa = 11.84QTGTYY166 pKa = 7.21KK167 pKa = 10.49TPVDD171 pKa = 3.64GTTGNDD177 pKa = 3.29SMGVGYY183 pKa = 9.23TDD185 pKa = 3.5AQGDD189 pKa = 4.06QITAGNDD196 pKa = 3.3YY197 pKa = 10.78ILAGAGNDD205 pKa = 3.37TVTAGDD211 pKa = 3.77GNDD214 pKa = 3.57FVSGGTGADD223 pKa = 4.33LIYY226 pKa = 10.74GGNGNDD232 pKa = 3.56SLQGNDD238 pKa = 4.27GNDD241 pKa = 3.51TIYY244 pKa = 11.43GEE246 pKa = 4.73LGADD250 pKa = 4.03TIDD253 pKa = 4.28GGAGDD258 pKa = 5.3DD259 pKa = 3.95SILYY263 pKa = 10.65GEE265 pKa = 4.89GADD268 pKa = 3.64VVYY271 pKa = 10.77GGAGNDD277 pKa = 3.88YY278 pKa = 10.79IDD280 pKa = 5.94DD281 pKa = 3.84INGAQLNGANTIYY294 pKa = 10.92GGDD297 pKa = 4.05GNDD300 pKa = 4.63SIYY303 pKa = 10.75TGNDD307 pKa = 2.63GDD309 pKa = 5.43LIYY312 pKa = 10.82GGQGTDD318 pKa = 3.34WIDD321 pKa = 3.61GEE323 pKa = 4.6LGNDD327 pKa = 3.52TVYY330 pKa = 11.24GGADD334 pKa = 3.54NDD336 pKa = 4.28TVMGGAGNDD345 pKa = 3.62SLFGGSGADD354 pKa = 3.94YY355 pKa = 10.84IDD357 pKa = 4.31GGADD361 pKa = 3.06NDD363 pKa = 4.51IIYY366 pKa = 10.88GDD368 pKa = 3.92FNGNTVNINYY378 pKa = 7.62TVSSSSLAASTPAARR393 pKa = 11.84VDD395 pKa = 3.91DD396 pKa = 4.64DD397 pKa = 3.82ATLKK401 pKa = 11.0VSFDD405 pKa = 3.27EE406 pKa = 4.51AATVRR411 pKa = 11.84LTFGLLEE418 pKa = 4.2NFEE421 pKa = 4.68EE422 pKa = 4.61NIISIDD428 pKa = 3.68GVVVDD433 pKa = 4.5IQAMINAGDD442 pKa = 3.74AVLSGGIAVGATGGVTGTAGSSTSGTITFNVPFTSISVDD481 pKa = 3.42HH482 pKa = 6.82TGGGYY487 pKa = 10.56DD488 pKa = 3.87DD489 pKa = 3.87VTVEE493 pKa = 3.95YY494 pKa = 10.01LATGGTYY501 pKa = 8.85TAITDD506 pKa = 3.9YY507 pKa = 11.82ANGATGSYY515 pKa = 10.47AADD518 pKa = 3.38TVGISGSDD526 pKa = 3.54TILGGLGDD534 pKa = 3.72DD535 pKa = 4.73TIWFGAGDD543 pKa = 3.95DD544 pKa = 3.89VVYY547 pKa = 10.83GGDD550 pKa = 3.9GNDD553 pKa = 4.49SIDD556 pKa = 3.81DD557 pKa = 3.72RR558 pKa = 11.84SGDD561 pKa = 3.66GLVGDD566 pKa = 4.77NYY568 pKa = 11.17LDD570 pKa = 4.02GGAGSDD576 pKa = 4.34TIWAGYY582 pKa = 10.48GSDD585 pKa = 3.55TVLGGDD591 pKa = 4.75GNDD594 pKa = 4.17SLHH597 pKa = 6.94GDD599 pKa = 3.46QGNDD603 pKa = 3.33TLSGGLGADD612 pKa = 4.14TLWGDD617 pKa = 3.83AGNDD621 pKa = 3.47VLDD624 pKa = 4.29GGEE627 pKa = 4.3GDD629 pKa = 3.83DD630 pKa = 4.79HH631 pKa = 7.52LVAGAGYY638 pKa = 8.02DD639 pKa = 3.69TIVASSGIDD648 pKa = 3.1NVYY651 pKa = 10.68QFDD654 pKa = 4.99LNDD657 pKa = 4.1DD658 pKa = 3.99NLDD661 pKa = 3.49GFTNHH666 pKa = 6.53QIDD669 pKa = 3.87VSNLTDD675 pKa = 4.24LEE677 pKa = 4.47GNPIRR682 pKa = 11.84WEE684 pKa = 4.08DD685 pKa = 3.56VVVTQNVNNHH695 pKa = 5.7AVLTFPNGEE704 pKa = 4.07QIILQGITADD714 pKa = 3.58QVNDD718 pKa = 3.63KK719 pKa = 11.22LEE721 pKa = 4.92LISIGIPCFTVGTMILTPTGEE742 pKa = 4.48KK743 pKa = 9.41PIEE746 pKa = 4.01MLRR749 pKa = 11.84PGDD752 pKa = 3.63MVVTRR757 pKa = 11.84DD758 pKa = 3.88NGPQPLVWAGSRR770 pKa = 11.84RR771 pKa = 11.84LGAGEE776 pKa = 4.29LARR779 pKa = 11.84HH780 pKa = 6.19PEE782 pKa = 3.85LRR784 pKa = 11.84PIRR787 pKa = 11.84IAPGDD792 pKa = 3.42WAGPRR797 pKa = 11.84GLLVSPQHH805 pKa = 6.29GLYY808 pKa = 10.34AHH810 pKa = 5.68QSEE813 pKa = 5.16RR814 pKa = 11.84GGTDD818 pKa = 2.87KK819 pKa = 11.26LIRR822 pKa = 11.84ATHH825 pKa = 6.41LARR828 pKa = 11.84LKK830 pKa = 10.16GGKK833 pKa = 9.19VRR835 pKa = 11.84VANGVTSVTYY845 pKa = 8.77IHH847 pKa = 6.98LMFEE851 pKa = 3.88EE852 pKa = 4.25HH853 pKa = 6.49QVIFGNGIASEE864 pKa = 4.21SFYY867 pKa = 10.34PGKK870 pKa = 9.93WGLSSLAAPCRR881 pKa = 11.84RR882 pKa = 11.84EE883 pKa = 3.6ILQLFPEE890 pKa = 4.59LAGTDD895 pKa = 3.42VEE897 pKa = 4.32NAYY900 pKa = 10.12GQTAMPFARR909 pKa = 11.84FKK911 pKa = 10.56EE912 pKa = 4.52LPDD915 pKa = 3.47HH916 pKa = 7.08VYY918 pKa = 10.95DD919 pKa = 6.05LSLQTFDD926 pKa = 5.48RR927 pKa = 11.84RR928 pKa = 11.84LLRR931 pKa = 11.84CGG933 pKa = 3.3
MM1 pKa = 7.14ATSTTFQVMYY11 pKa = 10.02LGQRR15 pKa = 11.84ALIDD19 pKa = 3.42TTQGNNTAEE28 pKa = 4.12NAAGILDD35 pKa = 4.34TYY37 pKa = 11.34GSASDD42 pKa = 4.76PMYY45 pKa = 10.83NWVGTLYY52 pKa = 10.99ADD54 pKa = 4.74YY55 pKa = 11.14LLEE58 pKa = 5.5DD59 pKa = 5.18DD60 pKa = 4.68NNSYY64 pKa = 11.11DD65 pKa = 3.35IDD67 pKa = 3.93NGGGRR72 pKa = 11.84DD73 pKa = 3.26RR74 pKa = 11.84FHH76 pKa = 8.1VNGFGTQEE84 pKa = 3.43FDD86 pKa = 3.89AVAEE90 pKa = 4.07YY91 pKa = 10.35AITLTYY97 pKa = 10.88VDD99 pKa = 4.32GTTAQITAYY108 pKa = 10.27VFQDD112 pKa = 3.76FQGHH116 pKa = 5.28TFLAPEE122 pKa = 4.17TTLNADD128 pKa = 3.49QAALTAKK135 pKa = 9.94PIQSLALTAVTKK147 pKa = 10.84SSGDD151 pKa = 3.39TTAGDD156 pKa = 4.08MIAARR161 pKa = 11.84QTGTYY166 pKa = 7.21KK167 pKa = 10.49TPVDD171 pKa = 3.64GTTGNDD177 pKa = 3.29SMGVGYY183 pKa = 9.23TDD185 pKa = 3.5AQGDD189 pKa = 4.06QITAGNDD196 pKa = 3.3YY197 pKa = 10.78ILAGAGNDD205 pKa = 3.37TVTAGDD211 pKa = 3.77GNDD214 pKa = 3.57FVSGGTGADD223 pKa = 4.33LIYY226 pKa = 10.74GGNGNDD232 pKa = 3.56SLQGNDD238 pKa = 4.27GNDD241 pKa = 3.51TIYY244 pKa = 11.43GEE246 pKa = 4.73LGADD250 pKa = 4.03TIDD253 pKa = 4.28GGAGDD258 pKa = 5.3DD259 pKa = 3.95SILYY263 pKa = 10.65GEE265 pKa = 4.89GADD268 pKa = 3.64VVYY271 pKa = 10.77GGAGNDD277 pKa = 3.88YY278 pKa = 10.79IDD280 pKa = 5.94DD281 pKa = 3.84INGAQLNGANTIYY294 pKa = 10.92GGDD297 pKa = 4.05GNDD300 pKa = 4.63SIYY303 pKa = 10.75TGNDD307 pKa = 2.63GDD309 pKa = 5.43LIYY312 pKa = 10.82GGQGTDD318 pKa = 3.34WIDD321 pKa = 3.61GEE323 pKa = 4.6LGNDD327 pKa = 3.52TVYY330 pKa = 11.24GGADD334 pKa = 3.54NDD336 pKa = 4.28TVMGGAGNDD345 pKa = 3.62SLFGGSGADD354 pKa = 3.94YY355 pKa = 10.84IDD357 pKa = 4.31GGADD361 pKa = 3.06NDD363 pKa = 4.51IIYY366 pKa = 10.88GDD368 pKa = 3.92FNGNTVNINYY378 pKa = 7.62TVSSSSLAASTPAARR393 pKa = 11.84VDD395 pKa = 3.91DD396 pKa = 4.64DD397 pKa = 3.82ATLKK401 pKa = 11.0VSFDD405 pKa = 3.27EE406 pKa = 4.51AATVRR411 pKa = 11.84LTFGLLEE418 pKa = 4.2NFEE421 pKa = 4.68EE422 pKa = 4.61NIISIDD428 pKa = 3.68GVVVDD433 pKa = 4.5IQAMINAGDD442 pKa = 3.74AVLSGGIAVGATGGVTGTAGSSTSGTITFNVPFTSISVDD481 pKa = 3.42HH482 pKa = 6.82TGGGYY487 pKa = 10.56DD488 pKa = 3.87DD489 pKa = 3.87VTVEE493 pKa = 3.95YY494 pKa = 10.01LATGGTYY501 pKa = 8.85TAITDD506 pKa = 3.9YY507 pKa = 11.82ANGATGSYY515 pKa = 10.47AADD518 pKa = 3.38TVGISGSDD526 pKa = 3.54TILGGLGDD534 pKa = 3.72DD535 pKa = 4.73TIWFGAGDD543 pKa = 3.95DD544 pKa = 3.89VVYY547 pKa = 10.83GGDD550 pKa = 3.9GNDD553 pKa = 4.49SIDD556 pKa = 3.81DD557 pKa = 3.72RR558 pKa = 11.84SGDD561 pKa = 3.66GLVGDD566 pKa = 4.77NYY568 pKa = 11.17LDD570 pKa = 4.02GGAGSDD576 pKa = 4.34TIWAGYY582 pKa = 10.48GSDD585 pKa = 3.55TVLGGDD591 pKa = 4.75GNDD594 pKa = 4.17SLHH597 pKa = 6.94GDD599 pKa = 3.46QGNDD603 pKa = 3.33TLSGGLGADD612 pKa = 4.14TLWGDD617 pKa = 3.83AGNDD621 pKa = 3.47VLDD624 pKa = 4.29GGEE627 pKa = 4.3GDD629 pKa = 3.83DD630 pKa = 4.79HH631 pKa = 7.52LVAGAGYY638 pKa = 8.02DD639 pKa = 3.69TIVASSGIDD648 pKa = 3.1NVYY651 pKa = 10.68QFDD654 pKa = 4.99LNDD657 pKa = 4.1DD658 pKa = 3.99NLDD661 pKa = 3.49GFTNHH666 pKa = 6.53QIDD669 pKa = 3.87VSNLTDD675 pKa = 4.24LEE677 pKa = 4.47GNPIRR682 pKa = 11.84WEE684 pKa = 4.08DD685 pKa = 3.56VVVTQNVNNHH695 pKa = 5.7AVLTFPNGEE704 pKa = 4.07QIILQGITADD714 pKa = 3.58QVNDD718 pKa = 3.63KK719 pKa = 11.22LEE721 pKa = 4.92LISIGIPCFTVGTMILTPTGEE742 pKa = 4.48KK743 pKa = 9.41PIEE746 pKa = 4.01MLRR749 pKa = 11.84PGDD752 pKa = 3.63MVVTRR757 pKa = 11.84DD758 pKa = 3.88NGPQPLVWAGSRR770 pKa = 11.84RR771 pKa = 11.84LGAGEE776 pKa = 4.29LARR779 pKa = 11.84HH780 pKa = 6.19PEE782 pKa = 3.85LRR784 pKa = 11.84PIRR787 pKa = 11.84IAPGDD792 pKa = 3.42WAGPRR797 pKa = 11.84GLLVSPQHH805 pKa = 6.29GLYY808 pKa = 10.34AHH810 pKa = 5.68QSEE813 pKa = 5.16RR814 pKa = 11.84GGTDD818 pKa = 2.87KK819 pKa = 11.26LIRR822 pKa = 11.84ATHH825 pKa = 6.41LARR828 pKa = 11.84LKK830 pKa = 10.16GGKK833 pKa = 9.19VRR835 pKa = 11.84VANGVTSVTYY845 pKa = 8.77IHH847 pKa = 6.98LMFEE851 pKa = 3.88EE852 pKa = 4.25HH853 pKa = 6.49QVIFGNGIASEE864 pKa = 4.21SFYY867 pKa = 10.34PGKK870 pKa = 9.93WGLSSLAAPCRR881 pKa = 11.84RR882 pKa = 11.84EE883 pKa = 3.6ILQLFPEE890 pKa = 4.59LAGTDD895 pKa = 3.42VEE897 pKa = 4.32NAYY900 pKa = 10.12GQTAMPFARR909 pKa = 11.84FKK911 pKa = 10.56EE912 pKa = 4.52LPDD915 pKa = 3.47HH916 pKa = 7.08VYY918 pKa = 10.95DD919 pKa = 6.05LSLQTFDD926 pKa = 5.48RR927 pKa = 11.84RR928 pKa = 11.84LLRR931 pKa = 11.84CGG933 pKa = 3.3
Molecular weight: 96.88 kDa
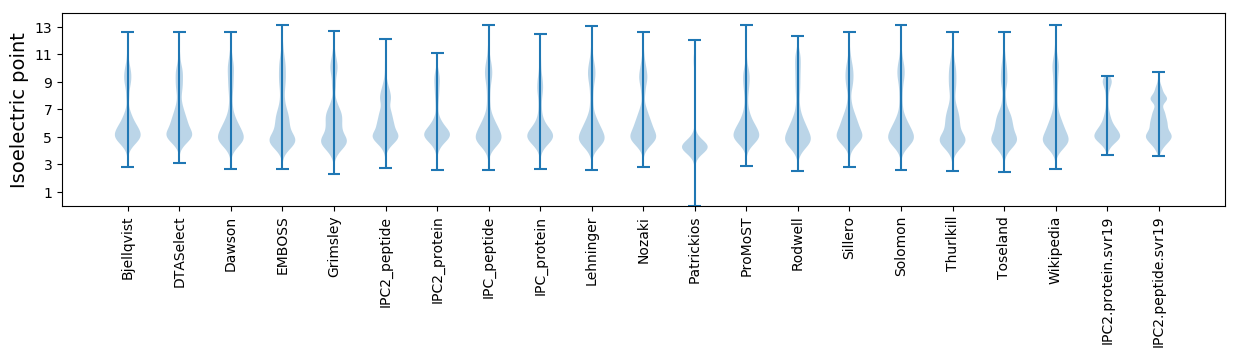
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2T0WZW2|A0A2T0WZW2_9RHOB Uncharacterized protein OS=Donghicola tyrosinivorans OX=1652492 GN=CLV74_102144 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.44RR3 pKa = 11.84TYY5 pKa = 10.31QPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37AGRR28 pKa = 11.84KK29 pKa = 8.54ILNARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.07SLSAA44 pKa = 3.93
MM1 pKa = 7.35KK2 pKa = 9.44RR3 pKa = 11.84TYY5 pKa = 10.31QPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37AGRR28 pKa = 11.84KK29 pKa = 8.54ILNARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.07SLSAA44 pKa = 3.93
Molecular weight: 5.15 kDa
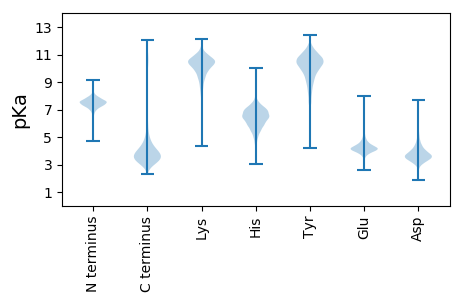
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1407917 |
26 |
18712 |
314.8 |
34.2 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.963 ± 0.046 | 0.919 ± 0.019 |
6.117 ± 0.043 | 5.867 ± 0.049 |
3.699 ± 0.031 | 8.567 ± 0.063 |
2.04 ± 0.026 | 5.353 ± 0.028 |
3.431 ± 0.039 | 9.983 ± 0.043 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.896 ± 0.028 | 2.857 ± 0.029 |
4.785 ± 0.05 | 3.438 ± 0.019 |
6.212 ± 0.056 | 5.314 ± 0.041 |
5.679 ± 0.084 | 7.231 ± 0.039 |
1.331 ± 0.016 | 2.318 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
